- Record: found
- Abstract: found
- Article: found
Role of pericytes in the development of cerebral cavernous malformations

Read this article at
Summary
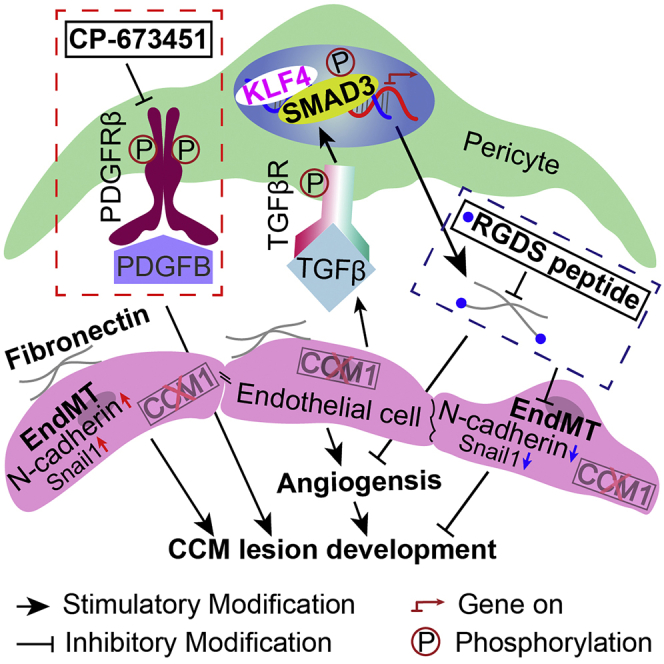
Cerebral cavernous malformation (CCM) is caused by loss-of-function mutations in CCM1, CCM2, or CCM3 genes of endothelial cells. It is characterized by pericyte deficiency. However, the role of pericytes in CCMs is not yet clarified. We found pericytes in Cdh5Cre ERT2 ; Ccm1 fl/fl ( Ccm1 ECKO ) mice had a high expression of PDGFRβ. The inhibition of pericyte function by CP-673451 aggravated the CCM lesion development. RNA-sequencing analysis revealed the molecular traits of pericytes, such as highly expressed ECM-related genes, especially Fn1. Furthermore, KLF4 coupled with phosphorylated SMAD3 (pSMAD3) promoted the transcription of fibronectin in the pericytes of CCM lesions. RGDS peptide, an inhibitor of fibronectin, decreased the lesion area in the cerebella and retinas of Ccm1 ECKO mice. Also, human CCM lesions had abundant fibronectin deposition, and pSMAD3- and KLF4-positive pericytes. These findings indicate that pericytes are essential for CCM lesion development, and fibronectin intervention may provide a novel target for therapeutic intervention in such patients.
Graphical abstract
Highlights
Abstract
Neuroscience; Neuroanatomy; Clinical neuroscience
Related collections
Most cited references91

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
clusterProfiler: an R package for comparing biological themes among gene clusters.

- Record: found
- Abstract: found
- Article: found