- Record: found
- Abstract: found
- Article: found
Early Life Stress and Epigenetics in Late-onset Alzheimer’s Dementia: A Systematic Review
Read this article at
Abstract
Involvement of life stress in Late-Onset Alzheimer’s Disease (LOAD) has been evinced in longitudinal cohort epidemiological studies, and endocrinologic evidence suggests involvements of catecholamine and corticosteroid systems in LOAD. Early Life Stress (ELS) rodent models have successfully demonstrated sequelae of maternal separation resulting in LOAD-analogous pathology, thereby supporting a role of insulin receptor signalling pertaining to GSK-3beta facilitated tau hyper-phosphorylation and amyloidogenic processing. Discussed are relevant ELS studies, and findings from three mitogen-activated protein kinase pathways (JNK/SAPK pathway, ERK pathway, p38/MAPK pathway) relevant for mediating environmental stresses. Further considered were the roles of autophagy impairment, neuroinflammation, and brain insulin resistance.
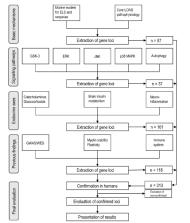
For the meta-analytic evaluation, 224 candidate gene loci were extracted from reviews of animal studies of LOAD pathophysiological mechanisms, of which 60 had no positive results in human LOAD association studies. These loci were combined with 89 gene loci confirmed as LOAD risk genes in previous GWAS and WES. Of the 313 risk gene loci evaluated, there were 35 human reports on epigenomic modifications in terms of methylation or histone acetylation. 64 microRNA gene regulation mechanisms were published for the compiled loci.
Genomic association studies support close relations of both noradrenergic and glucocorticoid systems with LOAD. For HPA involvement, a CRHR1 haplotype with MAPT was described, but further association of only HSD11B1 with LOAD found; however, association of FKBP1 and NC3R1 polymorphisms was documented in support of stress influence to LOAD. In the brain insulin system, IGF2R, INSR, INSRR, and plasticity regulator ARC, were associated with LOAD. Pertaining to compromised myelin stability in LOAD, relevant associations were found for BIN1, RELN, SORL1, SORCS1, CNP, MAG, and MOG. Regarding epigenetic modifications, both methylation variability and de-acetylation were reported for LOAD. The majority of up-to-date epigenomic findings include reported modifications in the well-known LOAD core pathology loci MAPT, BACE1, APP (with FOS, EGR1), PSEN1, PSEN2, and highlight a central role of BDNF. Pertaining to ELS, relevant loci are FKBP5, EGR1, GSK3B; critical roles of inflammation are indicated by CRP, TNFA, NFKB1 modifications; for cholesterol biosynthesis, DHCR24; for myelin stability BIN1, SORL1, CNP; pertaining to (epi)genetic mechanisms, hTERT, MBD2, DNMT1, MTHFR2. Findings on gene regulation were accumulated for BACE1, MAPK signalling, TLR4, BDNF, insulin signalling, with most reports for miR-132 and miR-27. Unclear in epigenomic studies remains the role of noradrenergic signalling, previously demonstrated by neuropathological findings of childhood nucleus caeruleus degeneration for LOAD tauopathy.
Related collections
Most cited references616
- Record: found
- Abstract: found
- Article: not found
Epigenetic programming by maternal behavior.
- Record: found
- Abstract: found
- Article: not found