- Record: found
- Abstract: found
- Article: found
Is FSH combined with equine chorionic gonadotropin able to modify lipid metabolism in bovine superstimulated antral follicles?
Abstract
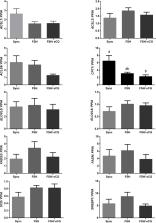
Lipid metabolism is essential for ensuring oocyte maturation and embryo development. β-Oxidized fatty acids (FA) are a potent source of energy for cells, particularly for bovine somatic follicular cells. Superstimulatory protocols using follicle stimulating hormone (FSH) or FSH combined with equine chorionic gonadotropin (eCG) are capable of stimulating the follicular microenvironment and drive the expression of biomarker genes associated with lipid metabolism in the cumulus-oocyte complex (COC) for better embryo development. In this study, we assesed the effects of FSH and FSH/eCG protocols on the expression of genes related to lipid metabolism in bovine granulosa cells (GCs). Further, we measured triglyceride levels in follicular fluid (FF) obtained from both superstimulatd and non-superstimulated cows (synchronized cows). In summary, superstimulation with gonadotropins maintained the TG levels in bovine FF and ensured GCs mRNA abundance of ACSL1, ACSL3, ACSL6, SCD, ELOVL5, ELOVL6, FASN, FADS2, and SREBP1. We, however, found the abundance of CPTIB mRNA to be lower in GCs obtained from cows subjected to FSH/eCG protocols than synchronized cows. In conclusion, the findings of this study showed that ovarian superstimulation around the preovulatory phase has a mild impact on the lipid metabolism in GCs.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
A new mathematical model for relative quantification in real-time RT-PCR.
- Record: found
- Abstract: found
- Article: not found
Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data.
