- Record: found
- Abstract: found
- Article: found
Rapid and Highly Stable Membrane Reconstitution by LAiR Enables the Study of Physiological Integral Membrane Protein Functions
Read this article at
Abstract

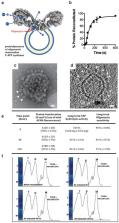
Functional reintegration into lipid environments represents a major challenge for in vitro investigation of integral membrane proteins (IMPs). Here, we report a new approach, termed LMNG Auto-insertion Reintegration (LAiR), for reintegration of IMPs into lipid bilayers within minutes. The resulting proteoliposomes displayed an unprecedented capability to maintain proton gradients and long-term stability. LAiR allowed for monitoring catalysis of a membrane-bound, physiologically relevant polyisoprenoid quinone substrate by Escherichia coli cytochromes bo 3 (c bo 3) and bd (c bd) under control of the proton motive force. LAiR also facilitated bulk-phase detection and physiological assessment of the “proton leak” in c bo 3, a controversial catalytic state that previously was only approachable at the single-molecule level. LAiR maintained the multisubunit integrity and higher-order oligomeric states of the delicate mammalian F-ATP synthase. Given that LAiR can be applied to both liposomes and planar membrane bilayers and is compatible with IMPs and lipids from prokaryotic and eukaryotic sources, we anticipate LAiR to be applied broadly across basic research, pharmaceutical applications, and biotechnology.
Abstract
LAiR: A low concentration LMNG based method for membrane auto-insertion, enabling rapid and stable lipid bilayer reconstitution of integral membrane proteins into liposomes and planar bilayers.
Related collections
Most cited references53
- Record: found
- Abstract: found
- Article: not found
Computer visualization of three-dimensional image data using IMOD.
- Record: found
- Abstract: found
- Article: not found
A comprehensive map of molecular drug targets
- Record: found
- Abstract: found
- Article: not found