- Record: found
- Abstract: found
- Article: found
Spatiotemporally-resolved mapping of RNA binding proteins via functional proximity labeling reveals a mitochondrial mRNA anchor promoting stress recovery
Read this article at
Abstract
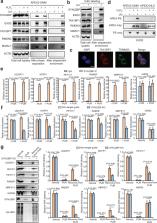
Proximity labeling (PL) with genetically-targeted promiscuous enzymes has emerged as a powerful tool for unbiased proteome discovery. By combining the spatiotemporal specificity of PL with methods for functional protein enrichment, we show that it is possible to map specific protein subclasses within distinct compartments of living cells. In particular, we develop a method to enrich subcompartment-specific RNA binding proteins (RBPs) by combining peroxidase-catalyzed PL with organic-aqueous phase separation of crosslinked protein-RNA complexes (“APEX-PS”). We use APEX-PS to generate datasets of nuclear, nucleolar, and outer mitochondrial membrane (OMM) RBPs, which can be mined for novel functions. For example, we find that the OMM RBP SYNJ2BP retains specific nuclear-encoded mitochondrial mRNAs at the OMM during translation stress, facilitating their local translation and import of protein products into the mitochondrion during stress recovery. Functional PL in general, and APEX-PS in particular, represent versatile approaches for the discovery of proteins with novel function in specific subcellular compartments.
Abstract
Proximity labeling is used to map and discover proteins in specific subcellular compartments. Here the authors combine APEX-mediated proximity labeling with organic-aqueous phase separation to identify nuclear, nucleolar, and outer mitochondrial membrane RNA binding proteins.
Related collections
Most cited references87
- Record: found
- Abstract: found
- Article: not found
A subcellular map of the human proteome
- Record: found
- Abstract: found
- Article: not found