- Record: found
- Abstract: found
- Article: found
Experimental insights into the importance of aquatic bacterial community composition to the degradation of dissolved organic matter
Read this article at
Abstract
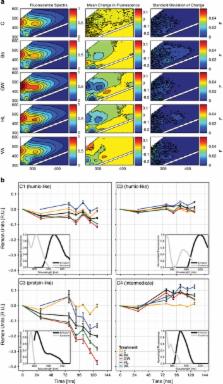
Bacteria play a central role in the cycling of carbon, yet our understanding of the relationship between the taxonomic composition and the degradation of dissolved organic matter (DOM) is still poor. In this experimental study, we were able to demonstrate a direct link between community composition and ecosystem functioning in that differently structured aquatic bacterial communities differed in their degradation of terrestrially derived DOM. Although the same amount of carbon was processed, both the temporal pattern of degradation and the compounds degraded differed among communities. We, moreover, uncovered that low-molecular-weight carbon was available to all communities for utilisation, whereas the ability to degrade carbon of greater molecular weight was a trait less widely distributed. Finally, whereas the degradation of either low- or high-molecular-weight carbon was not restricted to a single phylogenetic clade, our results illustrate that bacterial taxa of similar phylogenetic classification differed substantially in their association with the degradation of DOM compounds. Applying techniques that capture the diversity and complexity of both bacterial communities and DOM, our study provides new insight into how the structure of bacterial communities may affect processes of biogeochemical significance.
Related collections
Most cited references18

- Record: found
- Abstract: found
- Article: found
MTML-msBayes: Approximate Bayesian comparative phylogeographic inference from multiple taxa and multiple loci with rate heterogeneity
- Record: found
- Abstract: found
- Article: not found
Measurement of dissolved organic matter fluorescence in aquatic environments: an interlaboratory comparison.
- Record: found
- Abstract: found
- Article: not found