- Record: found
- Abstract: found
- Article: found
Molecular determinants of the interaction between HSV-1 glycoprotein D and heparan sulfate
Read this article at
Abstract
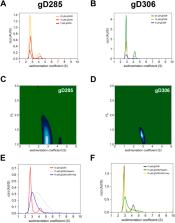
Literature has well-established the importance of 3- O-sulfation of neuronal cell surface glycan heparan sulfate (HS) to its interaction with herpes simplex virus type 1 glycoprotein D (gD). Previous investigations of gD to its viral receptors HVEM and nectin-1 also highlighted the conformational dynamics of gD’s N- and C-termini, necessary for viral membrane fusion. However, little is known on the structural interactions of gD with HS. Here, we present our findings on this interface from both the glycan and the protein perspective. We used C-terminal and N-terminal gD variants to probe the role of their respective regions in gD/HS binding. The N-terminal truncation mutants (with Δ1-22) demonstrate equivalent or stronger binding to heparin than their intact glycoproteins, indicating that the first 22 amino acids are disposable for heparin binding. Characterization of the conformational differences between C-terminal truncated mutants by sedimentation velocity analytical ultracentrifugation distinguished between the “open” and “closed” conformations of the glycoprotein D, highlighting the region’s modulation of receptor binding. From the glycan perspective, we investigated gD interacting with heparin, heparan sulfate, and other de-sulfated and chemically defined oligosaccharides using surface plasmon resonance and glycan microarray. The results show a strong preference of gD for 6- O-sulfate, with 2- O-sulfation becoming more important in the presence of 6- O-S. Additionally, 3- O-sulfation shifted the chain length preference of gD from longer chain to mid-chain length, reaffirming the sulfation site’s importance to the gD/HS interface. Our results shed new light on the molecular details of one of seven known protein-glycan interactions with 3- O-sulfated heparan sulfate.
Related collections
Most cited references73
- Record: found
- Abstract: found
- Article: not found
Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling.
- Record: found
- Abstract: found
- Article: not found
Heparan sulfate proteoglycans.
- Record: found
- Abstract: found
- Article: not found
Order out of chaos: assembly of ligand binding sites in heparan sulfate.
Author and article information
Comments
Comment on this article
 Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.