- Record: found
- Abstract: found
- Article: found
Genome-wide association study in individuals of European and African ancestry and multi-trait analysis of opioid use disorder identifies 19 independent genome-wide significant risk loci
Read this article at
Abstract
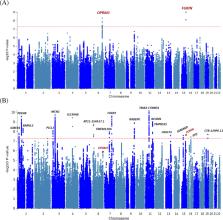
Despite the large toll of opioid use disorder (OUD), genome-wide association studies (GWAS) of OUD to date have yielded few susceptibility loci. We performed a large-scale GWAS of OUD in individuals of European (EUR) and African (AFR) ancestry, optimizing genetic informativeness by performing MTAG (Multi-trait analysis of GWAS) with genetically correlated substance use disorders (SUDs). Meta-analysis included seven cohorts: the Million Veteran Program, Psychiatric Genomics Consortium, iPSYCH, FinnGen, Partners Biobank, BioVU, and Yale-Penn 3, resulting in a total N = 639,063 ( N cases = 20,686;N effective = 77,026) across ancestries. OUD cases were defined as having a lifetime OUD diagnosis, and controls as anyone not known to meet OUD criteria. We estimated SNP-heritability (h 2 SNP) and genetic correlations (r g). Based on genetic correlation, we performed MTAG on OUD, alcohol use disorder (AUD), and cannabis use disorder (CanUD). A leave-one-out polygenic risk score (PRS) analysis was performed to compare OUD and OUD-MTAG PRS as predictors of OUD case status in Yale-Penn 3. The EUR meta-analysis identified three genome-wide significant (GWS; p ≤ 5 × 10 − 8) lead SNPs—one at FURIN (rs11372849; p = 9.54 × 10 − 10) and two OPRM1 variants (rs1799971, p = 4.92 × 10 − 09; rs79704991, p = 1.11 × 10 − 08; r 2 = 0.02). Rs1799971 (p = 4.91 × 10 − 08) and another OPRM1 variant (rs9478500; p = 1.95 × 10 − 08; r 2 = 0.03) were identified in the cross-ancestry meta-analysis. Estimated h 2 SNP was 12.75%, with strong r g with CanUD (r g = 0.82; p = 1.14 × 10 − 47) and AUD (r g = 0.77; p = 6.36 × 10 − 78). The OUD-MTAG resulted in a GWAS N equivalent = 128,748 and 18 independent GWS loci, some mapping to genes or gene regions that have previously been associated with psychiatric or addiction phenotypes. The OUD-MTAG PRS accounted for 3.81% of OUD variance (beta = 0.61;s.e. = 0.066; p = 2.00 × 10 − 16) compared to 2.41% (beta = 0.45; s.e. = 0.058; p = 2.90 × 10 − 13) explained by the OUD PRS. The current study identified OUD variant associations at OPRM1, single variant associations with FURIN, and 18 GWS associations in the OUD-MTAG. The genetic architecture of OUD is likely influenced by both OUD-specific loci and loci shared across SUDs.
Related collections
Most cited references43
- Record: found
- Abstract: not found
- Book: not found
Diagnostic and Statistical Manual of Mental Disorders

- Record: found
- Abstract: found
- Article: found
A global reference for human genetic variation
- Record: found
- Abstract: found
- Article: found