- Record: found
- Abstract: found
- Article: found
Discrepancies in tumor mutation burden reporting from sequential endobronchial ultrasound transbronchial needle aspiration samples within single lymph node stations - brief report
Read this article at
Abstract
Introduction
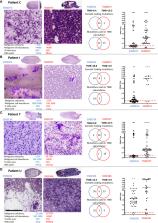
Tumour Mutation Burden (TMB) is a potential biomarker for immune cancer therapies. Here we investigated parameters that might affect TMB using duplicate cytology smears obtained from endobronchial ultrasound transbronchial needle aspiration (EBUS TBNA)-sampled malignant lymph nodes.
Methods
Individual Diff-Quik cytology smears were prepared for each needle pass. DNA extracted from each smear underwent sequencing using large gene panel (TruSight Oncology 500 (TSO500 - Illumina)). TMB was estimated using the TSO500 Local App v. 2.0 (Illumina).
Results
Twenty patients had two or more Diff-Quik smears (total 45 smears) which passed sequencing quality control. Average smear TMB was 8.7 ± 5.0 mutations per megabase (Mb). Sixteen of the 20 patients had paired samples with minimal differences in TMB score (average difference 1.3 ± 0.85). Paired samples from 13 patients had concordant TMB (scores below or above a threshold of 10 mutations/Mb). Markedly discrepant TMB was observed in four cases, with an average difference of 11.3 ± 2.7 mutations/Mb. Factors affecting TMB calling included sample tumour content, the amount of DNA used in sequencing, and bone fide heterogeneity of node tumour between paired samples.
Conclusion
TMB assessment is feasible from EBUS-TBNA smears from a single needle pass. Repeated samples of a lymph node station have minimal variation in TMB in most cases. However, this novel data shows how tumour content and minor change in site of node sampling can impact TMB. Further study is needed on whether all node aspirates should be combined in 1 sample, or whether testing independent nodes using smears is needed.
Related collections
Most cited references17
- Record: found
- Abstract: found
- Article: not found
Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer.
- Record: found
- Abstract: found
- Article: not found
Nivolumab plus Ipilimumab in Advanced Non–Small-Cell Lung Cancer
- Record: found
- Abstract: found
- Article: not found