- Record: found
- Abstract: found
- Article: found
Identification of a novel cathelicidin antimicrobial peptide from ducks and determination of its functional activity and antibacterial mechanism
Read this article at
Abstract
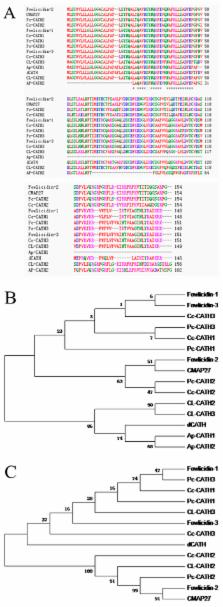
The family of antimicrobial peptide, cathelicidins, which plays important roles against infections in animals, has been identified from many species. Here, we identified a novel avian cathelicidin ortholog from ducks and named dCATH. The cDNA sequence of dCATH encodes a predicted 146-amino-acid polypeptide composed of a 17-residue signal peptide, a 109-residue conserved cathelin domain and a 20-residue mature peptide. Phylogenetic analysis demonstrated that dCATH is highly divergent from other avian peptides. The α-helical structure of the peptide exerted strong antimicrobial activity against a broad range of bacteria in vitro, with most minimum inhibitory concentrations in the range of 2 to 4 μM. Moreover, dCATH also showed cytotoxicity, lysing 50% of mammalian erythrocytes in the presence or absence of 10% fetal calf serum at concentrations of 32 μM or 20 μM and killing 50% HaCaT cells at a concentration of 10 μM. The effects on bacterial outer and inner membranes, as examined by scanning electron microscope and transmission electron microscopy, indicate that dCATH kills microbial cells by increasing permeability, causing a loss of membrane integrity.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3.
- Record: found
- Abstract: found
- Article: not found
Mechanism of action of the antimicrobial peptide buforin II: buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions.
- Record: found
- Abstract: found
- Article: not found