- Record: found
- Abstract: found
- Article: found
HAMSAB diet ameliorates dysfunctional signaling in pancreatic islets in autoimmune diabetes
Read this article at
Summary
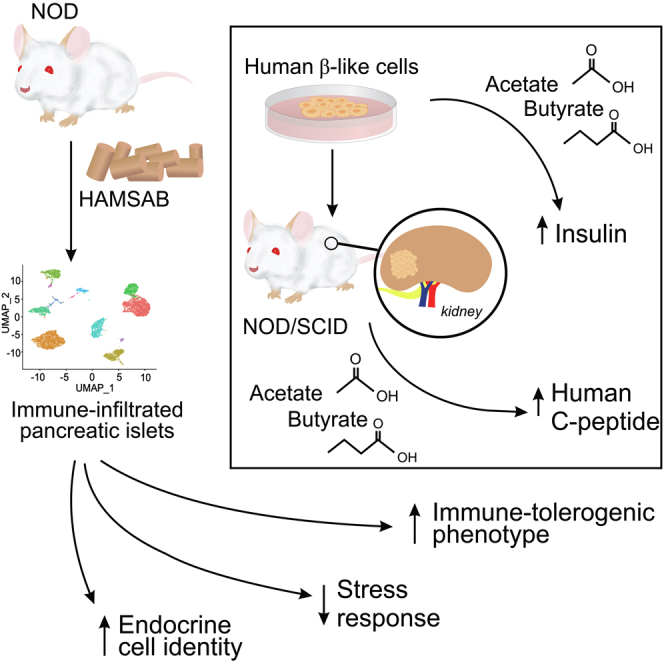
An altered gut microbiota is associated with type 1 diabetes (T1D), affecting the production of short-chain fatty acids (SCFA) and glucose homeostasis. We previously demonstrated that enhancing serum acetate and butyrate using a dietary supplement (HAMSAB) improved glycemia in non-obese diabetic (NOD) mice and patients with established T1D. The effects of SCFA on immune-infiltrated islet cells remain to be clarified. Here, we performed single-cell RNA sequencing on islet cells from NOD mice fed an HAMSAB or control diet. HAMSAB induced a regulatory gene expression profile in pancreas-infiltrated immune cells. Moreover, HAMSAB maintained the expression of β-cell functional genes and decreased cellular stress. HAMSAB-fed mice showed preserved pancreatic endocrine cell identity, evaluated by decreased numbers of poly-hormonal cells. Finally, SCFA increased insulin levels in human β-like cells and improved transplantation outcome in NOD/SCID mice. Our findings support the use of metabolite-based diet as attractive approach to improve glucose control in T1D.
Graphical abstract
Highlights
-
•
HAMSAB induces a tolerogenic phenotype of infiltrated immune cells in NOD mice
-
•
HAMSAB reduces the stress response maintaining endocrine cell identity in NOD mice
-
•
SCFAs increase insulin levels and function of human beta-like cells
-
•
HAMSAB is an attractive strategy for clinical studies in early type 1 diabetes
Abstract
Diabetology; Cell biology; Transcriptomics; Model organism
Related collections
Most cited references53
- Record: found
- Abstract: found
- Article: not found
Comprehensive Integration of Single-Cell Data

- Record: found
- Abstract: found
- Article: found
Integrated analysis of multimodal single-cell data

- Record: found
- Abstract: found
- Article: found