- Record: found
- Abstract: found
- Article: found
Performance of neural network basecalling tools for Oxford Nanopore sequencing

Read this article at
Abstract
Background
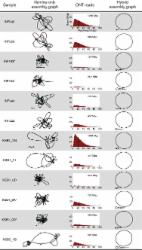
Basecalling, the computational process of translating raw electrical signal to nucleotide sequence, is of critical importance to the sequencing platforms produced by Oxford Nanopore Technologies (ONT). Here, we examine the performance of different basecalling tools, looking at accuracy at the level of bases within individual reads and at majority-rule consensus basecalls in an assembly. We also investigate some additional aspects of basecalling: training using a taxon-specific dataset, using a larger neural network model and improving consensus basecalls in an assembly by additional signal-level analysis with Nanopolish.
Results
Training basecallers on taxon-specific data results in a significant boost in consensus accuracy, mostly due to the reduction of errors in methylation motifs. A larger neural network is able to improve both read and consensus accuracy, but at a cost to speed. Improving consensus sequences (‘polishing’) with Nanopolish somewhat negates the accuracy differences in basecallers, but pre-polish accuracy does have an effect on post-polish accuracy.
Conclusions
Basecalling accuracy has seen significant improvements over the last 2 years. The current version of ONT’s Guppy basecaller performs well overall, with good accuracy and fast performance. If higher accuracy is required, users should consider producing a custom model using a larger neural network and/or training data from the same species.
Related collections
Most cited references19
- Record: found
- Abstract: found
- Article: found
Completing bacterial genome assemblies with multiplex MinION sequencing
- Record: found
- Abstract: found
- Article: not found
Messenger RNA modifications: Form, distribution, and function.

- Record: found
- Abstract: found
- Article: found