- Record: found
- Abstract: found
- Article: found
Emerging Evidence of Translational Control by AU-Rich Element-Binding Proteins
Read this article at
Abstract
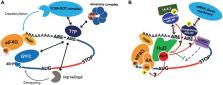
RNA-binding proteins (RBPs) are key regulators of posttranscriptional gene expression and control many important biological processes including cell proliferation, development, and differentiation. RBPs bind specific motifs in their target mRNAs and regulate mRNA fate at many steps. The AU-rich element (ARE) is one of the major cis-regulatory elements in the 3′ untranslated region (UTR) of labile mRNAs. Many of these encode factors requiring very tight regulation, such as inflammatory cytokines and growth factors. Disruption in the control of these factors’ expression can cause autoimmune diseases, developmental disorders, or cancers. Therefore, these mRNAs are strictly regulated by various RBPs, particularly ARE-binding proteins (ARE-BPs). To regulate mRNA metabolism, ARE-BPs bind target mRNAs and affect some factors on mRNAs directly, or recruit effectors, such as mRNA decay machinery and protein kinases to target mRNAs. Importantly, some ARE-BPs have stabilizing roles, whereas others are destabilizing, and ARE-BPs appear to compete with each other when binding to target mRNAs. The function of specific ARE-BPs is modulated by posttranslational modifications (PTMs) including methylation and phosphorylation, thereby providing a means for cellular signaling pathways to regulate stability of specific target mRNAs. In this review, we summarize recent studies which have revealed detailed molecular mechanisms of ARE-BP-mediated regulation of gene expression and also report on the importance of ARE-BP function in specific physiological contexts and how this relates to disease. We also propose an mRNP regulatory network based on competition between stabilizing ARE-BPs and destabilizing ARE-BPs.
Related collections
Most cited references97
- Record: found
- Abstract: found
- Article: not found
From birth to death: the complex lives of eukaryotic mRNAs.
- Record: found
- Abstract: found
- Article: not found
RNA recognition motifs: boring? Not quite.
- Record: found
- Abstract: found
- Article: not found