- Record: found
- Abstract: found
- Article: found
Insight into novel clinical mutants of RpsA-S324F, E325K, and G341R of Mycobacterium tuberculosis associated with pyrazinamide resistance
Read this article at
Abstract
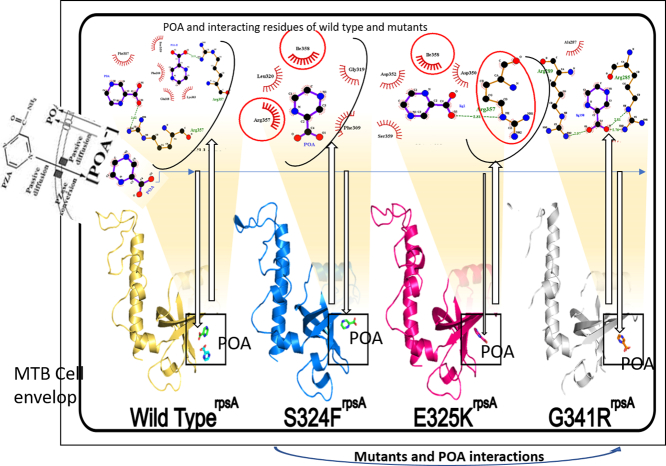
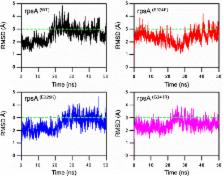
Pyrazinamide (PZA) is an important component of first-line anti-tuberculosis drugs which is converted into active form, pyrazinoic acid (POA), by Mycobacterium tuberculosis (MTB) pncA gene encoded, pyrazinamidase (PZase). Mutations in pncA are detected in >70% of PZA resistant isolates but, noticeably, not in all. In this study, we selected 18 PZA-resistant but wild type pncA (pncA WT) MTB isolates. Drug susceptibility testing (DST) of all the isolates were repeated at the critical concentration of PZA drug. All these PZA-resistance but pncA WT isolates were subjected to RpsA sequencing. Fifteen different mutations were identified in eleven isolates, where seven were present in a conserved region including, Ser324Phe, Glu325Lys, Gly341Arg. As the molecular mechanism of resistance behind these variants has not been reported earlier, we have performed multiple analysis to unveil the mechanisms of resistance behind mutations S324F, E325K, and G341R. The mutant and wild type RpsA structures were subjected to comprehensive computational molecular dynamic simulations at 50 ns. Root mean square deviation (RMSD), Root mean square fluctuation (RMSF), and Gibbs free energy of mutants were analyzed in comparison with wild type. Docking score of wild type -RpsA has been found to be maximum, showing a strong binding affinity in comparison with mutants. Pocket volume, RMSD and RMSF have also been found to be altered, whereas total energy, folding effect (radius of gyration) and shape complimentarily analysis showed that variants S324F, E325K, and G341R have been playing a significant role behind PZA-resistance. The study offers valuable information for better management of drug resistance tuberculosis.
Graphical abstract
Related collections
Most cited references33
- Record: found
- Abstract: not found
- Article: not found
Radius of gyration as an indicator of protein structure compactness
- Record: found
- Abstract: found
- Article: not found
Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery.
- Record: found
- Abstract: not found
- Article: not found