- Record: found
- Abstract: found
- Article: found
Molecular diversity and selective sweeps in maize inbred lines adapted to African highlands
Read this article at
Abstract
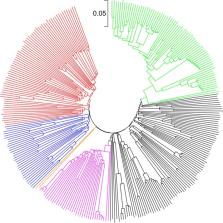
Little is known on maize germplasm adapted to the African highland agro-ecologies. In this study, we analyzed high-density genotyping by sequencing (GBS) data of 298 African highland adapted maize inbred lines to (i) assess the extent of genetic purity, genetic relatedness, and population structure, and (ii) identify genomic regions that have undergone selection (selective sweeps) in response to adaptation to highland environments. Nearly 91% of the pairs of inbred lines differed by 30–36% of the scored alleles, but only 32% of the pairs of the inbred lines had relative kinship coefficient <0.050, which suggests the presence of substantial redundancy in allelic composition that may be due to repeated use of fewer genetic backgrounds (source germplasm) during line development. Results from different genetic relatedness and population structure analyses revealed three different groups, which generally agrees with pedigree information and breeding history, but less so by heterotic groups and endosperm modification. We identified 944 single nucleotide polymorphic (SNP) markers that fell within 22 selective sweeps that harbored 265 protein-coding candidate genes of which some of the candidate genes had known functions. Details of the candidate genes with known functions and differences in nucleotide diversity among groups predicted based on multivariate methods have been discussed.
Related collections
Most cited references67
- Record: found
- Abstract: not found
- Article: not found
CDK inhibitors: positive and negative regulators of G1-phase progression.
- Record: found
- Abstract: found
- Article: not found
OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression.
- Record: found
- Abstract: found
- Article: not found