- Record: found
- Abstract: found
- Article: found
The clinical trials puzzle: How network effects limit drug discovery

Read this article at
Summary
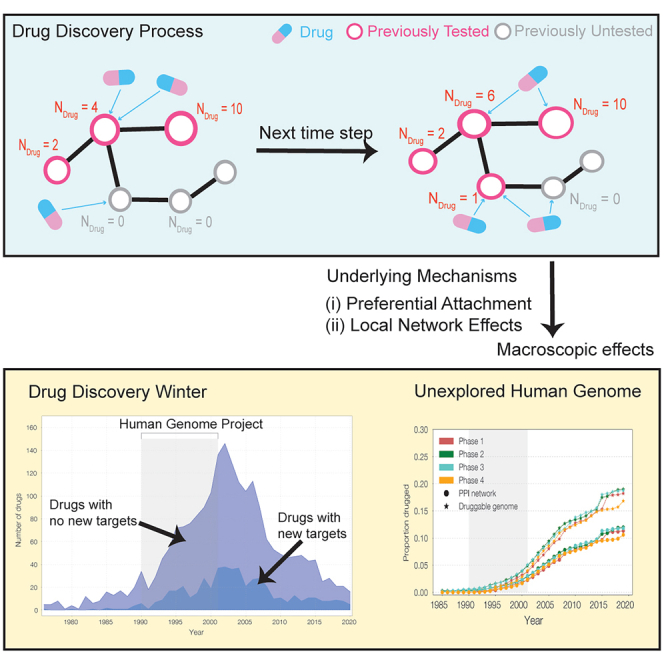
The depth of knowledge offered by post-genomic medicine has carried the promise of new drugs, and cures for multiple diseases. To explore the degree to which this capability has materialized, we extract meta-data from 356,403 clinical trials spanning four decades, aiming to offer mechanistic insights into the innovation practices in drug discovery. We find that convention dominates over innovation, as over 96% of the recorded trials focus on previously tested drug targets, and the tested drugs target only 12% of the human interactome. If current patterns persist, it would take 170 years to target all druggable proteins. We uncover two network-based fundamental mechanisms that currently limit target discovery: preferential attachment, leading to the repeated exploration of previously targeted proteins; and local network effects, limiting exploration to proteins interacting with highly explored proteins. We build on these insights to develop a quantitative network-based model to enhance drug discovery in clinical trials.
Graphical abstract
Highlights
Abstract
Medicine; Bioinformatics
Related collections
Most cited references45

- Record: found
- Abstract: found
- Article: found
DrugBank 5.0: a major update to the DrugBank database for 2018
- Record: found
- Abstract: found
- Article: not found
Emergence of Scaling in Random Networks

- Record: found
- Abstract: found
- Article: found