- Record: found
- Abstract: found
- Article: found
Circular RNAs in Diabetic Nephropathy: Updates and Perspectives
Read this article at
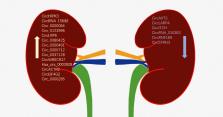
Abstract
Circular RNAs (circRNAs) are widespread endogenous transcripts lacking 5′-caps and 3′-polyadenylation tails. Their closed-loop structure confers exonuclease resistance and extreme stability. CircRNAs play essential roles in various diseases, including diabetes. Diabetic nephropathy (DN) is the leading cause of end-stage kidney disease and is one of the most common complications of diabetes. CircRNAs are key in DN and therefore important for understanding DN pathophysiology and developing new therapeutic strategies. In the present review, we briefly introduce the characteristics and functions of circRNAs and summarize recent discoveries on how circRNAs participate in DN. Based on these advances, we suggest future perspectives for studying circRNAs in DN to improve DN treatment and management.
Related collections
Most cited references119
- Record: found
- Abstract: found
- Article: not found
Natural RNA circles function as efficient microRNA sponges.
- Record: found
- Abstract: found
- Article: not found