- Record: found
- Abstract: found
- Article: found
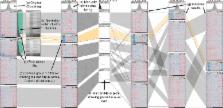
Interactive visual exploration and refinement of cluster assignments
Read this article at
Abstract
Background
With ever-increasing amounts of data produced in biology research, scientists are in need of efficient data analysis methods. Cluster analysis, combined with visualization of the results, is one such method that can be used to make sense of large data volumes. At the same time, cluster analysis is known to be imperfect and depends on the choice of algorithms, parameters, and distance measures. Most clustering algorithms don’t properly account for ambiguity in the source data, as records are often assigned to discrete clusters, even if an assignment is unclear. While there are metrics and visualization techniques that allow analysts to compare clusterings or to judge cluster quality, there is no comprehensive method that allows analysts to evaluate, compare, and refine cluster assignments based on the source data, derived scores, and contextual data.
Results
In this paper, we introduce a method that explicitly visualizes the quality of cluster assignments, allows comparisons of clustering results and enables analysts to manually curate and refine cluster assignments. Our methods are applicable to matrix data clustered with partitional, hierarchical, and fuzzy clustering algorithms. Furthermore, we enable analysts to explore clustering results in context of other data, for example, to observe whether a clustering of genomic data results in a meaningful differentiation in phenotypes.
Related collections
Most cited references24
- Record: found
- Abstract: not found
- Article: not found
FCM: The fuzzy c-means clustering algorithm
- Record: found
- Abstract: found
- Article: not found
D³: Data-Driven Documents.
- Record: found
- Abstract: not found
- Conference Proceedings: not found