- Record: found
- Abstract: found
- Article: not found
Regulatory association of long noncoding RNAs and chromatin accessibility facilitates erythroid differentiation

Read this article at
Key Points
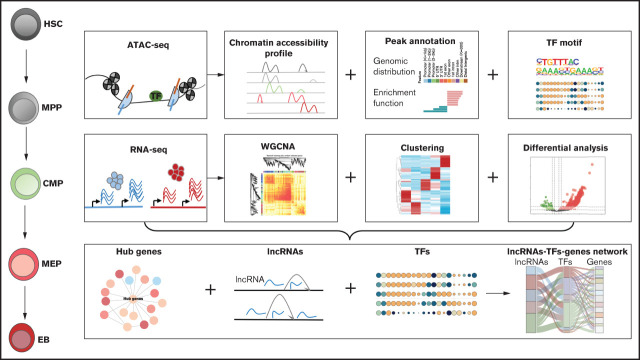
Visual Abstract
Abstract
Erythroid differentiation is a dynamic process regulated by multiple factors, whereas the interaction between long noncoding RNAs (lncRNAs) and chromatin accessibility and its influence on erythroid differentiation remains unclear. To elucidate this interaction, we used hematopoietic stem cells, multipotent progenitor cells, common myeloid progenitor cells, megakaryocyte-erythroid progenitor cells, and erythroblasts from human cord blood as an erythroid differentiation model to explore the coordinated regulatory functions of lncRNAs and chromatin accessibility by integrating RNA-seq and ATAC-seq data. We revealed that the integrated network of chromatin accessibility and lncRNAs exhibits stage-specific changes throughout the erythroid differentiation process and that the changes at the erythroblast stage of maturation are dramatic. We identified a subset of stage-specific lncRNAs and transcription factors (TFs) that associate with chromatin accessibility during erythroid differentiation, in which lncRNAs are key regulators of terminal erythroid differentiation via an lncRNA-TF-gene network. LncRNA PCED1B-AS1 was revealed to regulate terminal erythroid differentiation by coordinating GATA1 dynamically binding to the chromatin and interacting with the cytoskeleton network during erythroid differentiation. DANCR, another lncRNA that is highly expressed at the megakaryocyte-erythroid progenitor cell stage, was verified to promote erythroid differentiation by compromising megakaryocyte differentiation and coordinating with chromatin accessibility and TFs, such as RUNX1. Overall, our results identify the associated network of lncRNAs and chromatin accessibility in erythropoiesis and provide novel insights into erythroid differentiation and abundant resources for further study.
Related collections
Most cited references67
- Record: found
- Abstract: found
- Article: not found
STAR: ultrafast universal RNA-seq aligner.
- Record: found
- Abstract: found
- Article: not found
clusterProfiler: an R package for comparing biological themes among gene clusters.

- Record: found
- Abstract: found
- Article: found