- Record: found
- Abstract: found
- Article: found
A Biotinylated cpFIT-PNA Platform for the Facile Detection of Drug Resistance to Artemisinin in Plasmodium falciparum
Read this article at
Abstract

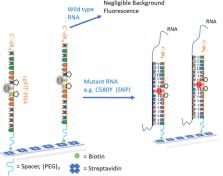
The evolution of drug resistance to many antimalarial drugs in the lethal strain of malaria ( Plasmodium falciparum) has been a great concern over the past 50 years. Among these drugs, artemisinin has become less effective for treating malaria. Indeed, several P. falciparum variants have become resistant to this drug, as elucidated by specific mutations in the pfK13 gene. This study presents the development of a diagnostic kit for the detection of a common point mutation in the pfK13 gene of P. falciparum, namely, the C580Y point mutation. FIT-PNAs (forced-intercalation peptide nucleic acid) are DNA mimics that serve as RNA sensors that fluoresce upon hybridization to their complementary RNA. Herein, FIT-PNAs were designed to sense the C580Y single nucleotide polymorphism (SNP) and were conjugated to biotin in order to bind these molecules to streptavidin-coated plates. Initial studies with synthetic RNA were conducted to optimize the sensing system. In addition, cyclopentane-modified PNA monomers (cpPNAs) were introduced to improve FIT-PNA sensing. Lastly, total RNA was isolated from red blood cells infected with P. falciparum (WT strain – NF54-WT or mutant strain – NF54-C580Y). Streptavidin plates loaded with either FIT-PNA or cpFIT-PNA were incubated with the total RNA. A significant difference in fluorescence for mutant vs WT total RNA was found only for the cpFIT-PNA probe. In summary, this study paves the way for a simple diagnostic kit for monitoring artemisinin drug resistance that may be easily adapted to malaria endemic regions.
Related collections
Most cited references63
- Record: found
- Abstract: found
- Article: not found
A molecular marker of artemisinin-resistant Plasmodium falciparum malaria.
- Record: found
- Abstract: found
- Article: not found
PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules.
- Record: found
- Abstract: found
- Article: not found