- Record: found
- Abstract: found
- Article: found
The genome of Eimeria falciformis - reduction and specialization in a single host apicomplexan parasite
Read this article at
Abstract
Background
The phylum Apicomplexa comprises important unicellular human parasites such as Toxoplasma and Plasmodium. Eimeria is the largest and most diverse genus of apicomplexan parasites and some species of the genus are the causative agent of coccidiosis, a disease economically devastating in poultry. We report a complete genome sequence of the mouse parasite Eimeria falciformis. We assembled and annotated the genome sequence to study host-parasite interactions in this understudied genus in a model organism host.
Results
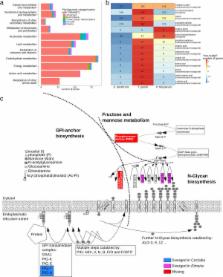
The genome of E. falciformis is 44 Mb in size and contains 5,879 predicted protein coding genes. Comparative analysis of E. falciformis with Toxoplasma gondii shows an emergence and diversification of gene families associated with motility and invasion mainly at the level of the Coccidia. Many rhoptry kinases, among them important virulence factors in T. gondii, are absent from the E. falciformis genome. Surface antigens are divergent between Eimeria species. Comparisons with T. gondii showed differences between genes involved in metabolism, N-glycan and GPI-anchor synthesis. E. falciformis possesses a reduced set of transmembrane transporters and we suggest an altered mode of iron uptake in the genus Eimeria.
Conclusions
Reduced diversity of genes required for host-parasite interaction and transmembrane transport allow hypotheses on host adaptation and specialization of a single host parasite. The E. falciformis genome sequence sheds light on the evolution of the Coccidia and helps to identify determinants of host-parasite interaction critical for drug and vaccine development.
Related collections
Most cited references57
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology
- Record: found
- Abstract: found
- Article: not found
MRBAYES: Bayesian inference of phylogenetic trees.
- Record: found
- Abstract: not found
- Article: not found