- Record: found
- Abstract: found
- Article: found
Genomic epidemiology of CVA10 in Guangdong, China, 2013–2021

Read this article at
Abstract
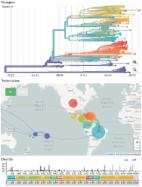
Hand, Foot and Mouth Disease (HFMD) is a highly contagious viral illness primarily affecting children globally. A significant epidemiological transition has been noted in mainland China, characterized by a substantial increase in HFMD cases caused by non-Enterovirus A71 (EV-A71) and non-Coxsackievirus A16 (CVA16) enteroviruses (EVs). Our study conducts a retrospective examination of 36,461 EV-positive specimens collected from Guangdong, China, from 2013 to 2021. Epidemiological trends suggest that, following 2013, Coxsackievirus A6 (CVA6) and Coxsackievirus A10 (CVA10) have emerged as the primary etiological agents for HFMD. In stark contrast, the incidence of EV-A71 has sharply declined, nearing extinction after 2018. Notably, cases of CVA10 infection were considerably younger, with a median age of 1.8 years, compared to 2.3 years for those with EV-A71 infections, possibly indicating accumulated EV-A71-specific herd immunity among young children. Through extensive genomic sequencing and analysis, we identified the N136D mutation in the 2 A protein, contributing to a predominant subcluster within genogroup C of CVA10 circulating in Guangdong since 2017. Additionally, a high frequency of recombination events was observed in genogroup F of CVA10, suggesting that the prevalence of this lineage might be underrecognized. The dynamic landscape of EV genotypes, along with their potential to cause outbreaks, underscores the need to broaden surveillance efforts to include a more diverse spectrum of EV genotypes. Moreover, given the shifting dominance of EV genotypes, it may be prudent to re-evaluate and optimize existing vaccination strategies, which are currently focused primarily target EV-A71.
Related collections
Most cited references31

- Record: found
- Abstract: found
- Article: found
MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability

- Record: found
- Abstract: found
- Article: found
IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies
- Record: found
- Abstract: found
- Article: found