- Record: found
- Abstract: found
- Article: found
Thyroid hormone signaling specifies cone subtypes in human retinal organoids

Read this article at
Abstract
INTRODUCTION:
Cone photoreceptors in the human retina enable daytime, color, and high-acuity vision. The three subtypes of human cones are defined by the visual pigment that they express: blue-opsin (short wavelength; S), green-opsin (medium wavelength; M), or red-opsin (long wavelength; L). Mutations that affect opsin expression or function cause various forms of color blindness and retinal degeneration.
RATIONALE:
Our current understanding of the vertebrate eye has been derived primarily from the study of model organisms. We studied the human retina to understand the developmental mechanisms that generate the mosaic of mutually exclusive cone subtypes. Specification of human cones occurs in a two-step process. First, a decision occurs between S versus L/M cone fates. If the L/M fate is chosen, a subsequent choice is made between expression of L- or M-opsin. To determine the mechanism that controls the first decision between S and L/M cone fates, we studied human retinal organoids derived from stem cells.
RESULTS:
We found that human organoids and retinas have similar distributions, gene expression profiles, and morphologies of cone subtypes. During development, S cones are specified first, followed by L/M cones. This temporal switch from specification of S cones to generation of L/M cones is controlled by thyroid hormone (TH) signaling. In retinal organoids that lacked thyroid hormone receptor β ( Thrβ), all cones developed into the S subtype. Thrβ binds with high affinity to triiodothyronine (T3), the more active form of TH, to regulate gene expression. We observed that addition of T3 early during development induced L/M fate in nearly all cones. Thus, TH signaling through Thrβ is necessary and sufficient to induce L/M cone fate and suppress S fate. TH exists largely in two states: thyroxine (T4), the most abundant circulating form of TH, and T3, which binds TH receptors with high affinity. We hypothesized that the retina itself could modulate TH levels to control subtype fates. We found that deiodinase 3 ( DIO3), an enzyme that degrades both T3 and T4, was expressed early in organoid and retina development. Conversely, deiodinase 2 ( DIO2), an enzyme that converts T4 to active T3, as well as TH carriers and transporters, were expressed later in development. Temporally dynamic expression of TH-degrading and -activating proteins supports a model in which the retina itself controls TH levels, ensuring low TH signaling early to specify S cones and high TH signaling later in development to produce L/M cones.
CONCLUSION:
Studies of model organisms and human epidemiology often generate hypotheses about human biology that cannot be studied in humans. Organoids provide a system to determine the mechanisms of human development, enabling direct testing of hypotheses in developing human tissue. Our studies identify temporal regulation of TH signaling as a mechanism that controls cone subtype specification in humans. Consistent with our findings, preterm human infants with low T3 and T4 have an increased incidence of color vision defects. Moreover, our identification of a mechanism that generates one cone subtype while suppressing the other, coupled with successful transplantation and incorporation of stem cell-derived photoreceptors in mice, suggests that the promise of therapies to treat human diseases such as color blindness, retinitis pigmentosa, and macular degeneration will be achieved in the near future. ■
Graphical Abstract
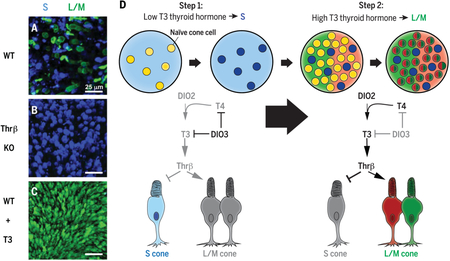
Temporally regulated TH signaling specifies cone subtypes. ( A) Embryonic stem cell-derived human retinal organoids [wild type (WT)] generate S and L/M cones. Blue, S-opsin; green, L/M-opsin. ( B) Organoids that lack thyroid hormone receptor β ( Thrβ KO) generate all S cones. ( C) Early activation of TH signaling (WT + T3) specifies nearly all L/M cones. ( D) TH-degrading enzymes (such as DIO3) expressed early in development lower TH and promote S fate, whereas TH-activating regulators (such as DIO2) expressed later promote L/M fate.
Summary
The mechanisms underlying specification of neuronal subtypes within the human nervous system are largely unknown. The blue (S), green (M), and red (L) cones of the retina enable high-acuity daytime and color vision. To determine the mechanism that controls S versus L/M fates, we studied the differentiation of human retinal organoids. Organoids and retinas have similar distributions, expression profiles, and morphologies of cone subtypes. S cones are specified first, followed by L/M cones, and thyroid hormone signaling controls this temporal switch. Dynamic expression of thyroid hormone–degrading and –activating proteins within the retina ensures low signaling early to specify S cones and high signaling late to produce L/M cones. This work establishes organoids as a model for determining mechanisms of human development with promising utility for therapeutics and vision repair.
Related collections
Most cited references48
- Record: found
- Abstract: found
- Article: not found
Biochemistry, cellular and molecular biology, and physiological roles of the iodothyronine selenodeiodinases.
- Record: found
- Abstract: found
- Article: not found
Transplantation of human embryonic stem cell-derived photoreceptors restores some visual function in Crx-deficient mice.
- Record: found
- Abstract: found
- Article: not found