- Record: found
- Abstract: found
- Article: not found
Orm family proteins mediate sphingolipid homeostasis
Read this article at
Abstract
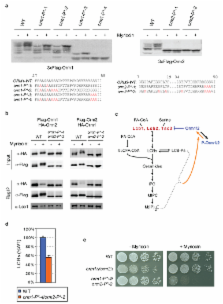
Despite the essential roles of sphingolipids as both structural components of membranes and critical signalling molecules, we have a limited understanding of how cells sense and regulate their levels. Here we reveal the function in sphingolipid metabolism of the ORM/ORMDL genes, a conserved gene family that includes ORMDL3, which has recently been identified as a potential risk factor for childhood asthma. Starting from an unbiased functional genomic approach, we identify Orm proteins as negative regulators of sphingolipid synthesis that form a conserved complex with serine palmitoyltransferase, the first and rate-limiting enzyme in sphingolipid production. We also define a regulatory pathway in which phosphorylation of Orm proteins relieves their inhibitory activity when sphingolipid production is disrupted. Changes in ORM gene expression or mutations to their phosphorylation sites cause dysregulation of sphingolipid metabolism. Our work identifies the Orm proteins as critical mediators of sphingolipid homeostasis and raises the possibility that sphingolipid misregulation contributes to the development of childhood asthma.
Related collections
Most cited references43
- Record: found
- Abstract: found
- Article: not found
Phosphate-binding tag, a new tool to visualize phosphorylated proteins.
- Record: found
- Abstract: found
- Article: not found
Molecular machinery for non-vesicular trafficking of ceramide.
- Record: found
- Abstract: found
- Article: not found
