- Record: found
- Abstract: found
- Article: found
Site-specific gene expression profiling as a novel strategy for unravelling keloid disease pathobiology
Read this article at
Abstract
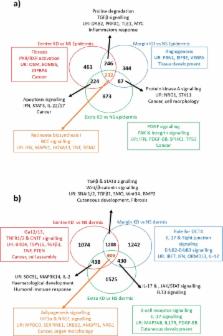
Keloid disease (KD) is a fibroproliferative cutaneous tumour characterised by heterogeneity, excess collagen deposition and aggressive local invasion. Lack of a validated animal model and resistance to a multitude of current therapies has resulted in unsatisfactory clinical outcomes of KD management. In order to address KD from a new perspective, we applied for the first time a site-specific in situ microdissection and gene expression profiling approach, through combined laser capture microdissection and transcriptomic array. The aim here was to analyse the utility of this approach compared with established methods of investigation, including whole tissue biopsy and monolayer cell culture techniques. This study was designed to approach KD from a hypothesis-free and compartment-specific angle, using state-of-the-art microdissection and gene expression profiling technology. We sought to characterise expression differences between specific keloid lesional sites and elucidate potential contributions of significantly dysregulated genes to mechanisms underlying keloid pathobiology, thus informing future explorative research into KD. Here, we highlight the advantages of our in situ microdissection strategy in generating expression data with improved sensitivity and accuracy over traditional methods. This methodological approach supports an active role for the epidermis in the pathogenesis of KD through identification of genes and upstream regulators implicated in epithelial-mesenchymal transition, inflammation and immune modulation. We describe dermal expression patterns crucial to collagen deposition that are associated with TGFβ-mediated signalling, which have not previously been examined in KD. Additionally, this study supports the previously proposed presence of a cancer-like stem cell population in KD and explores the possible contribution of gene dysregulation to the resistance of KD to conventional therapy. Through this innovative in situ microdissection gene profiling approach, we provide better-defined gene signatures of distinct KD regions, thereby addressing KD heterogeneity, facilitating differential diagnosis with other cutaneous fibroses via transcriptional fingerprinting, and highlighting key areas for future KD research.
Related collections
Most cited references150
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing
- Record: found
- Abstract: not found
- Article: not found
What is principal component analysis?
- Record: found
- Abstract: found
- Article: not found