- Record: found
- Abstract: found
- Article: found
Rhizosphere protists are key determinants of plant health
Read this article at
Abstract
Background
Plant health is intimately influenced by the rhizosphere microbiome, a complex assembly of organisms that changes markedly across plant growth. However, most rhizosphere microbiome research has focused on fractions of this microbiome, particularly bacteria and fungi. It remains unknown how other microbial components, especially key microbiome predators—protists—are linked to plant health. Here, we investigated the holistic rhizosphere microbiome including bacteria, microbial eukaryotes (fungi and protists), as well as functional microbial metabolism genes. We investigated these communities and functional genes throughout the growth of tomato plants that either developed disease symptoms or remained healthy under field conditions.
Results
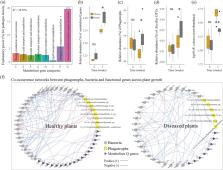
We found that pathogen dynamics across plant growth is best predicted by protists. More specifically, communities of microbial-feeding phagotrophic protists differed between later healthy and diseased plants at plant establishment. The relative abundance of these phagotrophs negatively correlated with pathogen abundance across plant growth, suggesting that predator-prey interactions influence pathogen performance. Furthermore, phagotrophic protists likely shifted bacterial functioning by enhancing pathogen-suppressing secondary metabolite genes involved in mitigating pathogen success.
Related collections
Most cited references39
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing

- Record: found
- Abstract: found
- Article: found
Soil networks become more connected and take up more carbon as nature restoration progresses
- Record: found
- Abstract: found
- Article: not found