- Record: found
- Abstract: found
- Article: found
Evolutionary activation of acidic chitinase in herbivores through the H128R mutation in ruminant livestock

Read this article at
Summary
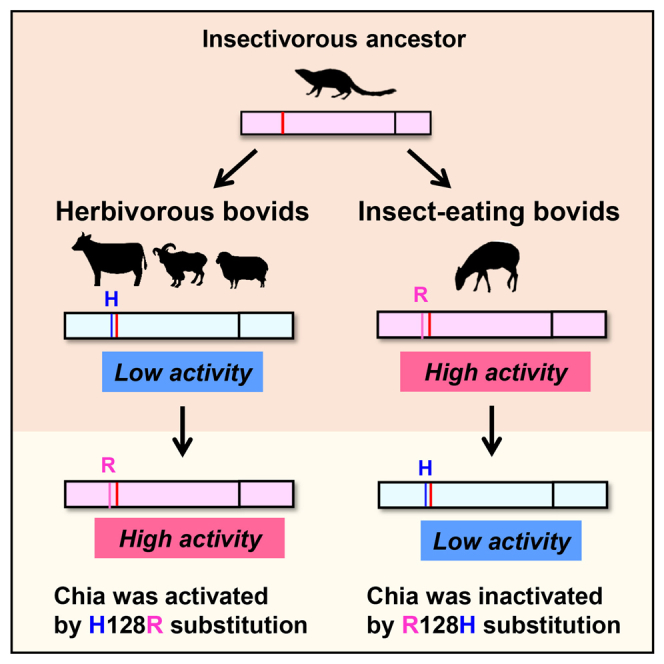
Placental mammals' ancestors were insectivores, suggesting that modern mammals may have inherited the ability to digest insects. Acidic chitinase (Chia) is a crucial enzyme hydrolyzing significant component of insects' exoskeleton in many species. On the other hand, herbivorous animal groups, such as cattle, have extremely low chitinase activity compared to omnivorous species, e.g., mice. The low activity of cattle Chia has been attributed to R128H mutation. The presence of either of these amino acids correlates with the feeding behavior of different bovid species with R and H determining the high and low enzymatic activity, respectively. Evolutionary analysis indicated that selective constraints were relaxed in 67 herbivorous Chia in Cetartiodactyla. Despite searching for another Chia paralog that could compensate for the reduced chitinase activity, no active paralogs were found in this order. Herbivorous animals' Chia underwent genetic alterations and evolved into a molecule with low activity due to the chitin-free diet.
Graphical abstract
Highlights
-
•
Herbivorous cattle have low chitinase activity due to R128H mutation
-
•
Insect-eating bovids have high chitinase activity due to the retention of R128
-
•
No active Chia paralogs were found in herbivorous Cetartiodactyla species
-
•
Selective constraints were relaxed in 67 herbivorous Chia genes in Cetartiodactyla
Abstract
Evolutionary biology; Molecular biology; Zoology.
Related collections
Most cited references51
- Record: found
- Abstract: found
- Article: not found
MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms.

- Record: found
- Abstract: found
- Article: found
Highly accurate protein structure prediction with AlphaFold
- Record: found
- Abstract: found
- Article: not found