- Record: found
- Abstract: found
- Article: found
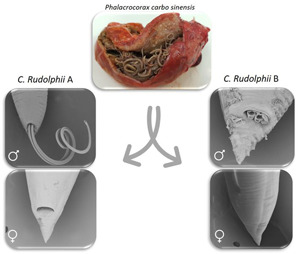
Molecular and morphological studies on Contracaecum rudolphii A and C. rudolphii B in great cormorants ( Phalacrocorax carbo sinensis) from Italy and Israel

Read this article at
Abstract
Abstract
The distribution of parasites is shaped by a variety of factors, among which are the migratory movements of their hosts. Israel has a unique position to migratory routes of several bird species leaving Europe to winter in Africa, however, detailed studies on the parasite fauna of birds from this area are scarce. Our study investigates occurrence and distribution of sibling species among Contracaecum rudolphii complex in Phalacrocorax carbo sinensis from Italy and Israel, to acquire further information on the geographical range of these species to gain deeper knowledge on the ecology of these parasites and their bird host. A total of 2383 Contracaecum were collected from the gastric mucosa of 28 great cormorants (18 from Israel and 10 from Italy). A subsample was processed for morphological analyses in light and scanning electron microscopy (SEM), and for molecular analyses through amplification and sequencing of the ITS rDNA and the cox2 mtDNA, and through PCR-RFLP. All the 683 Contracaecum subjected to molecular identification belonged to C. rudolphii s.l., (300 C. rudolphii A and 383 C. rudolphii B). SEM micrographs provided, for the first time, details of taxonomic structures in male specimens from both sibling species, and the first SEM characterization of C. rudolphii B. This work presents the first data on the occurrence of sibling species of C. rudolphii in Israel and provides additional information on the distribution of C. rudolphii A and B in Italy, confirming the high prevalence and intensity of infection observed in Ph. carbo sinensis from other Italian areas.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: not found
MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets.
- Record: found
- Abstract: found
- Article: found
MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space
- Record: found
- Abstract: found
- Article: not found