- Record: found
- Abstract: found
- Article: not found
Home chemical and microbial transitions across urbanization
Read this article at
Abstract
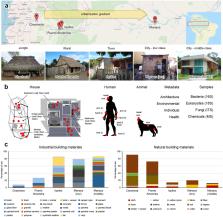
Urbanization represents a profound shift in human behavior, with significant cultural and health-associated consequences 2, 3 . Here we investigate chemical and microbial characteristics of houses and their human occupants across an urbanization gradient in the Amazon rainforest, from a remote Peruvian Amerindian village to the Brazilian city of Manaus. Urbanization was associated with reduced microbial outdoor exposure, increased contact with housing materials, antimicrobials, and cleaning products, and increased exposure to chemical diversity. Urbanization degree correlated with changes in house bacterial and micro-eukaryotic community composition, increased house and skin fungal diversity, and increased relative abundance of human skin-associated fungi and bacteria in houses. Overall, our results indicate large-scale effects of urbanization on chemical and microbial exposures and on the human microbiota.
Related collections
Most cited references44
- Record: found
- Abstract: found
- Article: not found
Cytoscape: a software environment for integrated models of biomolecular interaction networks.

- Record: found
- Abstract: found
- Article: found
The SILVA ribosomal RNA gene database project: improved data processing and web-based tools
- Record: found
- Abstract: not found
- Article: not found
