- Record: found
- Abstract: found
- Article: found
Cistrome and transcriptome analysis identifies unique androgen receptor (AR) and AR-V7 splice variant chromatin binding and transcriptional activities
Read this article at
Abstract
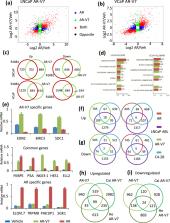
The constitutively active androgen receptor (AR) splice variant, AR-V7, plays an important role in resistance to androgen deprivation therapy in castration resistant prostate cancer (CRPC). Studies seeking to determine whether AR-V7 is a partial mimic of the AR, or also has unique activities, and whether the AR-V7 cistrome contains unique binding sites have yielded conflicting results. One limitation in many studies has been the low level of AR variant compared to AR. Here, LNCaP and VCaP cell lines in which AR-V7 expression can be induced to match the level of AR, were used to compare the activities of AR and AR-V7. The two AR isoforms shared many targets, but overall had distinct transcriptomes. Optimal induction of novel targets sometimes required more receptor isoform than classical targets such as PSA. The isoforms displayed remarkably different cistromes with numerous differential binding sites. Some of the unique AR-V7 sites were located proximal to the transcription start sites (TSS). A de novo binding motif similar to a half ARE was identified in many AR-V7 preferential sites and, in contrast to conventional half ARE sites that bind AR-V7, FOXA1 was not enriched at these sites. This supports the concept that the AR isoforms have unique actions with the potential to serve as biomarkers or novel therapeutic targets.
Related collections
Most cited references59
- Record: found
- Abstract: found
- Article: not found
STAR: ultrafast universal RNA-seq aligner.
- Record: found
- Abstract: found
- Article: not found
Fast gapped-read alignment with Bowtie 2.

- Record: found
- Abstract: found
- Article: found