- Record: found
- Abstract: found
- Article: found
Clinical genetic testing using a custom-designed steroid-resistant nephrotic syndrome gene panel: analysis and recommendations
Read this article at
Abstract
Background
There are many single-gene causes of steroid-resistant nephrotic syndrome (SRNS) and the list continues to grow rapidly. Prompt comprehensive diagnostic testing is key to realising the clinical benefits of a genetic diagnosis. This report describes a bespoke-designed, targeted next-generation sequencing (NGS) diagnostic gene panel assay to detect variants in 37 genes including the ability to identify copy number variants (CNVs).
Methods
This study reports results of 302 patients referred for SRNS diagnostic gene panel analysis. Phenotype and clinical impact data were collected using a standard proforma. Candidate variants detected by NGS were confirmed by Sanger sequencing/Multiplex Ligation-dependent Probe Amplification with subsequent family segregation analysis where possible.
Results
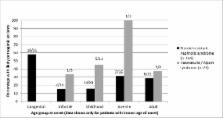
Clinical presentation was nephrotic syndrome in 267 patients and suspected Alport syndrome (AS) in 35. NGS panel testing determined a likely genetic cause of disease in 44/220 (20.0%) paediatric and 10/47 (21.3%) adult nephrotic cases, and 17/35 (48.6%) of haematuria/AS patients. Of 71 patients with genetic disease, 32 had novel pathogenic variants without a previous disease association including two with deletions of one or more exons of NPHS1 or NPHS2.
Conclusion
Gene panel testing provides a genetic diagnosis in a significant number of patients presenting with SRNS or suspected AS. It should be undertaken at an early stage of the care pathway and include the ability to detect CNVs as an emerging mechanism for genes associated with this condition. Use of clinical genetic testing after diagnosis of SRNS has the potential to stratify patients and assist decision-making regarding management.
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness.
- Record: found
- Abstract: found
- Article: not found
Spectrum of steroid-resistant and congenital nephrotic syndrome in children: the PodoNet registry cohort.
- Record: found
- Abstract: found
- Article: not found