- Record: found
- Abstract: found
- Article: found
The m 5C methyltransferase NSUN2 promotes codon‐dependent oncogenic translation by stabilising tRNA in anaplastic thyroid cancer
Read this article at
Abstract
Background
Translation dysregulation plays a crucial role in tumourigenesis and cancer progression. Oncogenic translation relies on the stability and availability of tRNAs for protein synthesis, making them potential targets for cancer therapy.
Methods
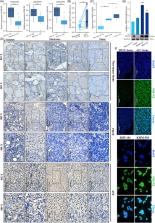
This study performed immunohistochemistry analysis to assess NSUN2 levels in thyroid cancer. Furthermore, to elucidate the impact of NSUN2 on anaplastic thyroid cancer (ATC) malignancy, phenotypic assays were conducted. Drug inhibition and time‐dependent plots were employed to analyse drug resistance. Liquid chromatography–mass spectrometry and bisulphite sequencing were used to investigate the m 5C methylation of tRNA at both global and single‐base levels. Puromycin intake and high‐frequency codon reporter assays verified the protein translation level. By combining mRNA and ribosome profiling, a series of downstream proteins and codon usage bias were identified. The acquired data were further validated by tRNA sequencing.
Results
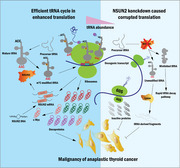
This study observed that the tRNA m 5C methyltransferase NSUN2 was up‐regulated in ATC and is associated with dedifferentiation. Furthermore, NSUN2 knockdown repressed ATC formation, proliferation, invasion and migration both in vivo and in vitro. Moreover, NSUN2 repression enhanced the sensitivity of ATC to genotoxic drugs. Mechanically, NSUN2 catalyses tRNA structure‐related m 5C modification, stabilising tRNA that maintains homeostasis and rapidly transports amino acids, particularly leucine. This stable tRNA has a substantially increased efficiency necessary to support a pro‐cancer translation program including c‐Myc, BCL2, RAB31, JUNB and TRAF2. Additionally, the NSUN2‐mediated variations in m5C levels and different tRNA Leu iso‐decoder families, partially contribute to a codon‐dependent translation bias. Surprisingly, targeting NSUN2 disrupted the c‐Myc to NSUN2 cycle in ATC.
Conclusions
This research revealed that a pro‐tumour m5C methyltransferase, dynamic tRNA stability regulation and downstream oncogenes, c‐Myc, elicits a codon‐dependent oncogenic translation network that enhances ATC growth and formation. Furthermore, it provides new opportunities for targeting translation reprogramming in cancer cells.
Abstract
In brief
P. Li et al. reports the m 5C methyltransferase NSUN2 catalyzes tRNA m 5C modification and regulates tRNA stability that is required for oncoprotein translation reprogramming and vicious dedifferentiation in anaplastic thyroid cancer cells. The study provides new opportunities for targetingcodon‐dependent oncogenic translation network.
Highlights
1. NSUN2 extensively catalyzes the tRNA m 5C modification, which is associated with tRNA secondary structure.
2. NSUN2 regulates tRNA stability more than aminoacylation to support selectivecodon‐dependent oncoprotein synthesis.
3. NSUN2 promotes maturation of its upstream transcription factor, c‐Myc.
4. Targeting NSUN2 exhibited broad anti‐cancer effects in vitro and in vivo.
5. NSUN2 maintains drug resistance in cancer cells.
Related collections
Most cited references104
- Record: found
- Abstract: found
- Article: not found
The 'effective number of codons' used in a gene.
- Record: found
- Abstract: found
- Article: not found
SUnSET, a nonradioactive method to monitor protein synthesis.
- Record: found
- Abstract: found
- Article: not found