- Record: found
- Abstract: found
- Article: not found
Cytoplasmic localization of alteration/deficiency in activation 3 (ADA3) predicts poor clinical outcome in breast cancer patients
Read this article at
Abstract
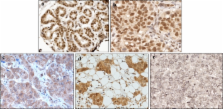
Transcriptional activation by estrogen receptor (ER) is a key step to breast oncogenesis. Given previous findings that ADA3 is a critical component of HAT complexes that regulate ER function and evidence that overexpression of other ER coactivators such as SRC-3 is associated with clinical outcomes in breast cancer, the current study was designed to assess the potential significance of ADA3 expression/localization in human breast cancer patients. In this study, we analyzed ADA3 expression in breast cancer tissue specimens and assessed the correlation of ADA3 staining with cancer progression and patient outcome. Tissue microarrays prepared from large series of breast cancer patients with long-term follow-ups were stained with anti-ADA3 monoclonal antibody using immunohistochemistry. Samples were analyzed for ADA3 expression followed by correlation with various clinicopathological parameters and patients’ outcomes. We report that breast cancer specimens show predominant nuclear, cytoplasmic, or mixed nuclear + cytoplasmic ADA3 staining patterns. Predominant nuclear ADA3 staining correlated with ER+ status. While predominant cytoplasmic ADA3 staining negatively correlated with ER+ status, but positively correlated with ErbB2, EGFR, and Ki67. Furthermore, a positive correlation of cytoplasmic ADA3 was observed with higher histological grade, mitotic counts, Nottingham Prognostic Index, and positive vascular invasion. Patients with nuclear ADA3 and ER positivity have better breast cancer specific survival and distant metastasis free survival. Significantly, cytoplasmic expression of ADA3 showed a strong positive association with reduced BCSS and DMFS in ErbB2+/EGFR+ patients. Although in multivariate analyses ADA3 expression was not an independent marker of survival, predominant nuclear ADA3 staining in breast cancer tissues correlates with ER+ expression and together serves as a marker of good prognosis, whereas predominant cytoplasmic ADA3 expression correlates with ErbB2+/EGFR+ expression and together is a marker of poor prognosis. Thus, ADA3 cytoplasmic localization together with ErbB2+/EGFR+ status may serve as better prognostic marker than individual proteins to predict survival of patients.
Related collections
Most cited references34
- Record: found
- Abstract: found
- Article: not found
A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1.
- Record: found
- Abstract: found
- Article: not found
Histone acetyltransferase complexes: one size doesn't fit all.
- Record: found
- Abstract: found
- Article: not found
