- Record: found
- Abstract: found
- Article: found
Enhancing of chemical compound and drug name recognition using representative tag scheme and fine-grained tokenization

Read this article at
Abstract
Background
The functions of chemical compounds and drugs that affect biological processes and their particular effect on the onset and treatment of diseases have attracted increasing interest with the advancement of research in the life sciences. To extract knowledge from the extensive literatures on such compounds and drugs, the organizers of BioCreative IV administered the CHEMical Compound and Drug Named Entity Recognition (CHEMDNER) task to establish a standard dataset for evaluating state-of-the-art chemical entity recognition methods.
Methods
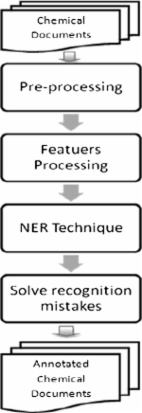
This study introduces the approach of our CHEMDNER system. Instead of emphasizing the development of novel feature sets for machine learning, this study investigates the effect of various tag schemes on the recognition of the names of chemicals and drugs by using conditional random fields. Experiments were conducted using combinations of different tokenization strategies and tag schemes to investigate the effects of tag set selection and tokenization method on the CHEMDNER task.
Results
This study presents the performance of CHEMDNER of three more representative tag schemes-IOBE, IOBES, and IOB 12E-when applied to a widely utilized IOB tag set and combined with the coarse-/fine-grained tokenization methods. The experimental results thus reveal that the fine-grained tokenization strategy performance best in terms of precision, recall and F-scores when the IOBES tag set was utilized. The IOBES model with fine-grained tokenization yielded the best-F-scores in the six chemical entity categories other than the "Multiple" entity category. Nonetheless, no significant improvement was observed when a more representative tag schemes was used with the coarse or fine-grained tokenization rules. The best F-scores that were achieved using the developed system on the test dataset of the CHEMDNER task were 0.833 and 0.815 for the chemical documents indexing and the chemical entity mention recognition tasks, respectively.
Conclusions
The results herein highlight the importance of tag set selection and the use of different tokenization strategies. Fine-grained tokenization combined with the tag set IOBES most effectively recognizes chemical and drug names. To the best of the authors' knowledge, this investigation is the first comprehensive investigation use of various tag set schemes combined with different tokenization strategies for the recognition of chemical entities.
Related collections
Most cited references6
- Record: found
- Abstract: found
- Article: not found
ChemSpot: a hybrid system for chemical named entity recognition.

- Record: found
- Abstract: found
- Article: found
Detection of IUPAC and IUPAC-like chemical names
- Record: found
- Abstract: found
- Article: found