- Record: found
- Abstract: found
- Article: found
Overexpression of Hevea brasiliensis ethylene response factor HbERF‐IXc5 enhances growth and tolerance to abiotic stress and affects laticifer differentiation
Read this article at
Summary
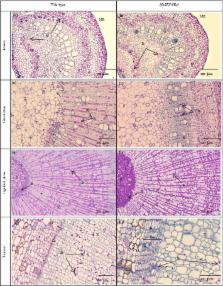
Ethylene response factor 1 ( ERF1) is an essential integrator of the jasmonate and ethylene signalling pathways coordinating a large number of genes involved in plant defences. Its orthologue in Hevea brasiliensis, Hb ERF‐ IXc5 , has been assumed to play a major role in laticifer metabolism and tolerance to harvesting stress for better latex production. This study sets out to establish and characterize rubber transgenic lines overexpressing Hb ERF‐ IXc5 . Overexpression of Hb ERF‐ IXc5 dramatically enhanced plant growth and enabled plants to maintain some ecophysiological parameters in response to abiotic stress such as water deficit, cold and salt treatments. This study revealed that Hb ERF‐ IXc5 has rubber‐specific functions compared to Arabidopsis ERF1 as transgenic plants overexpressing Hb ERF‐ IXc5 accumulated more starch and differentiated more latex cells at the histological level. The role of Hb ERF‐ IXc5 in driving the expression of some target genes involved in laticifer differentiation is discussed.
Related collections
Most cited references65
- Record: found
- Abstract: found
- Article: not found
Some relationships between the biochemistry of photosynthesis and the gas exchange of leaves.
- Record: found
- Abstract: found
- Article: not found
Genome-wide analysis of the ERF gene family in Arabidopsis and rice.
- Record: found
- Abstract: found
- Article: not found