- Record: found
- Abstract: found
- Article: found
Towards the Segmentation and Classification of White Blood Cell Cancer Using Hybrid Mask-Recurrent Neural Network and Transfer Learning
Read this article at
Abstract
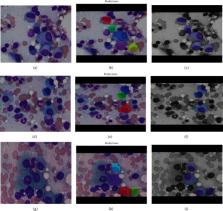
Inside the bone marrow, plasma cells are created, and they are a type of white blood cells. They are made from B lymphocytes. Antigens are produced by plasma cells to combat bacteria and viruses and prevent inflammation and illness. Multiple myeloma is a plasma cell cancer that starts in the bone marrow and causes the formation of abnormal plasma cells. Multiple myeloma is firmly identified by examining bone marrow samples under a microscope for myeloma cells. To diagnose myeloma cells, pathologists have to be very selective. Furthermore, because the ultimate decision is based on human sight and opinion, there is a possibility of error in the result. The nobility of this research is that it provides a computer-assisted technique for recognizing and detecting myeloma cells in bone marrow smears. For recognizing purposes, we have used Mask-Recurrent Convolutional Neural Network, and for detection purposes, Efficient Net B3 has been used. There are already many studies on white blood cell cancer, but very few with both segmentation and classification. We have designed two models. One is for recognizing myeloma cells, and the other is for differentiating them from nonmyeloma cells. Also, a new data set has been made from the multiple myeloma data sets, which has been used in our classification model. This research focuses on hybrid segmentation models and increases the accuracy level of the classification model. Both of our models are trained pretty well, where the Mask-RCNN model gives a mean average precision (mAP) of 93% and the Efficient Net B3 model gives 94.68% accuracy. The result of this research indicates that the Mask-RCNN model can recognize multiple myeloma and Efficient Net B3 can distinguish between myeloma and nonmyeloma cells and beats most of the state of the art in myeloma recognition and detection.
Related collections
Most cited references26
- Record: found
- Abstract: not found
- Conference Proceedings: not found
Deep Residual Learning for Image Recognition
- Record: found
- Abstract: found
- Article: found
Deep Learning in Medical Image Analysis

- Record: found
- Abstract: found
- Article: found