- Record: found
- Abstract: found
- Article: found
Metabolic Maturation Media Improve Physiological Function of Human iPSC-Derived Cardiomyocytes
Read this article at
SUMMARY
Induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) have enormous potential for the study of human cardiac disorders. However, their physiological immaturity severely limits their utility as a model system and their adoption for drug discovery. Here, we describe maturation media designed to provide oxidative substrates adapted to the metabolic needs of human iPSC (hiPSC)-CMs. Compared with conventionally cultured hiPSC-CMs, metabolically matured hiPSC-CMs contract with greater force and show an increased reliance on cardiac sodium (Na +) channels and sarcoplasmic reticulum calcium (Ca 2+) cycling. The media enhance the function, long-term survival, and sarcomere structures in engineered heart tissues. Use of the maturation media made it possible to reliably model two genetic cardiac diseases: long QT syndrome type 3 due to a mutation in the cardiac Na + channel SCN5A and dilated cardiomyopathy due to a mutation in the RNA splicing factor RBM20. The maturation media should increase the fidelity of hiPSC-CMs as disease models.
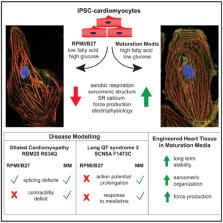
Graphical Abstract

In Brief
Physiological immaturity of iPSC-derived cardiomyocytes limits their fidelity as disease models. Feyen et al. developed a low glucose, high oxidative substrate media that increase maturation of ventricular-like hiPSC-CMs in 2D and 3D cultures relative to standard protocols. Improved characteristics include a low resting V m, rapid depolarization, and increased Ca 2+ dependence and force generation.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
Cardiac metabolism and its interactions with contraction, growth, and survival of cardiomyocytes.
- Record: found
- Abstract: found
- Article: not found
Thyroid and Glucocorticoid Hormones Promote Functional T-Tubule Development in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes.

- Record: found
- Abstract: found
- Article: found