- Record: found
- Abstract: found
- Article: found
Pathogenic Mechanisms in Centronuclear Myopathies
Read this article at
Abstract
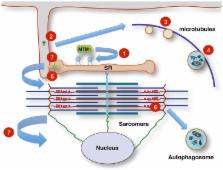
Centronuclear myopathies (CNMs) are a genetically heterogeneous group of inherited neuromuscular disorders characterized by clinical features of a congenital myopathy and abundant central nuclei as the most prominent histopathological feature. The most common forms of congenital myopathies with central nuclei have been attributed to X-linked recessive mutations in the MTM1 gene encoding myotubularin (“X-linked myotubular myopathy”), autosomal-dominant mutations in the DNM2 gene encoding dynamin-2 and the BIN1 gene encoding amphiphysin-2 (also named bridging integrator-1, BIN1, or SH3P9), and autosomal-recessive mutations in BIN1, the RYR1 gene encoding the skeletal muscle ryanodine receptor, and the TTN gene encoding titin. Models to study and rescue the affected cellular pathways are now available in yeast, C. elegans, drosophila, zebrafish, mouse, and dog. Defects in membrane trafficking have emerged as a key pathogenic mechanisms, with aberrant T-tubule formation, abnormalities of triadic assembly, and disturbance of the excitation–contraction machinery the main downstream effects studied to date. Abnormal autophagy has recently been recognized as another important collateral of defective membrane trafficking in different genetic forms of CNM, suggesting an intriguing link to primary disorders of defective autophagy with overlapping histopathological features. The following review will provide an overview of clinical, histopathological, and genetic aspects of the CNMs in the context of the key pathogenic mechanism, outline unresolved questions, and indicate promising future lines of enquiry.
Related collections
Most cited references113
- Record: found
- Abstract: found
- Article: not found
BAR domains as sensors of membrane curvature: the amphiphysin BAR structure.
- Record: found
- Abstract: found
- Article: not found
The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond.
- Record: found
- Abstract: found
- Article: not found