- Record: found
- Abstract: found
- Article: found
Oncogenic Gain of Function in Glioblastoma Is Linked to Mutant p53 Amyloid Oligomers

Read this article at
Summary
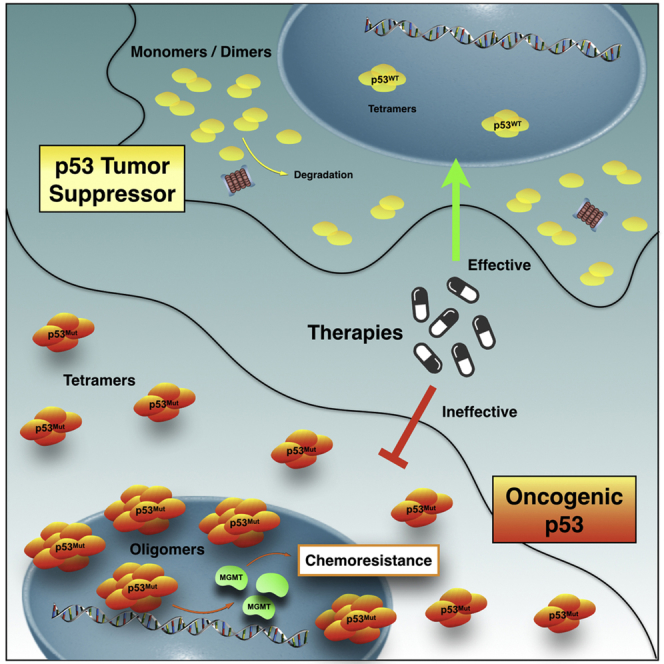
Tumor-associated p53 mutations endow cells with malignant phenotypes, including chemoresistance. Amyloid-like oligomers of mutant p53 transform this tumor suppressor into an oncogene. However, the composition and distribution of mutant p53 oligomers are unknown and the mechanism involved in the conversion is sparse. Here, we report accumulation of a p53 mutant within amyloid-like p53 oligomers in glioblastoma-derived cells presenting a chemoresistant gain-of-function phenotype. Statistical analysis from fluorescence fluctuation spectroscopy, pressure-induced measurements, and thioflavin T kinetics demonstrates the distribution of oligomers larger than the active tetrameric form of p53 in the nuclei of living cells and the destabilization of native-drifted p53 species that become amyloid. Collectively, these results provide insights into the role of amyloid-like mutant p53 oligomers in the chemoresistance phenotype of malignant and invasive brain tumors and shed light on therapeutic options to avert cancer.
Graphical Abstract
Highlights
Abstract
Structural Biology; Protein Structure Aspects; Biophysics; Cancer
Related collections
Most cited references57
- Record: found
- Abstract: found
- Article: not found
DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2.
- Record: found
- Abstract: found
- Article: found
Genetic pathways to primary and secondary glioblastoma.
- Record: found
- Abstract: found
- Article: not found