- Record: found
- Abstract: found
- Article: found
Broad anti-pathogen potential of DEAD box RNA helicase eIF4A-targeting rocaglates
Read this article at
Abstract
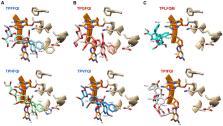
Inhibition of eukaryotic initiation factor 4A has been proposed as a strategy to fight pathogens. Rocaglates exhibit the highest specificities among eIF4A inhibitors, but their anti-pathogenic potential has not been comprehensively assessed across eukaryotes. In silico analysis of the substitution patterns of six eIF4A1 aa residues critical to rocaglate binding, uncovered 35 variants. Molecular docking of eIF4A:RNA:rocaglate complexes, and in vitro thermal shift assays with select recombinantly expressed eIF4A variants, revealed that sensitivity correlated with low inferred binding energies and high melting temperature shifts. In vitro testing with silvestrol validated predicted resistance in Caenorhabditis elegans and Leishmania amazonensis and predicted sensitivity in Aedes sp., Schistosoma mansoni, Trypanosoma brucei, Plasmodium falciparum, and Toxoplasma gondii. Our analysis further revealed the possibility of targeting important insect, plant, animal, and human pathogens with rocaglates. Finally, our findings might help design novel synthetic rocaglate derivatives or alternative eIF4A inhibitors to fight pathogens.
Related collections
Most cited references62
- Record: found
- Abstract: found
- Article: not found
AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading.
- Record: found
- Abstract: found
- Article: not found
AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility.
- Record: found
- Abstract: found
- Article: not found