- Record: found
- Abstract: found
- Article: found
Redox regulation by TXNRD3 during epididymal maturation underlies capacitation-associated mitochondrial activity and sperm motility in mice
Read this article at
Abstract
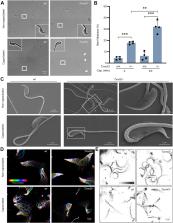
During epididymal transit, redox remodeling protects mammalian spermatozoa, preparing them for survival in the subsequent journey to fertilization. However, molecular mechanisms of redox regulation in sperm development and maturation remain largely elusive. In this study, we report that thioredoxin-glutathione reductase (TXNRD3), a thioredoxin reductase family member particularly abundant in elongating spermatids at the site of mitochondrial sheath formation, regulates redox homeostasis to support male fertility. Using Txnrd3 −/− mice, our biochemical, ultrastructural, and live cell imaging analyses revealed impairments in sperm morphology and motility under conditions of TXNRD3 deficiency. We find that mitochondria develop more defined cristae during capacitation in wildtype sperm. Furthermore, we show that absence of TXNRD3 alters thiol redox status in both the head and tail during sperm maturation and capacitation, resulting in defective mitochondrial ultrastructure and activity under capacitating conditions. These findings provide insights into molecular mechanisms of redox homeostasis and bioenergetics during sperm maturation, capacitation, and fertilization.
Related collections
Most cited references72
- Record: found
- Abstract: found
- Article: not found
Fiji: an open-source platform for biological-image analysis.
- Record: found
- Abstract: found
- Article: not found
Comprehensive Integration of Single-Cell Data
- Record: found
- Abstract: found
- Article: not found