- Record: found
- Abstract: found
- Article: not found
Longitudinal Changes in the Gut Microbiome of Infants on Total Parenteral Nutrition
Read this article at
Abstract
Background:
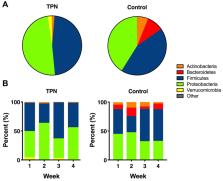
Animal studies suggest that total parenteral nutrition (TPN) may alter bacterial colonization of the intestinal tract and contribute to complications. Progressive changes in gut microbiome of infants receiving TPN are not well understood.
Methods:
Infants with and without TPN/soy lipid were enrolled in a prospective, longitudinal study. Weekly fecal samples were obtained for the first 4 weeks of life. High throughput pyrosequencing of 16S rDNA was used for compositional analysis of the gut microbiome.
Results:
47 infants were eligible for analyses, 25 infants received TPN and 22 infants did not (control). Although similar between TPN and control groups in the first week, fecal bacterial alpha diversity was significantly lower in the TPN group compared to controls at week 4 (Shannon index 1.0 vs 1.5, P-value = 0.03). The TPN group had significantly lower Bacteroidetes and higher Verrucomicrobia abundance compared to controls (P-values <0.05), and these differences became more pronounced over time. At the genus level, TPN was associated with lower abundance of Bacteroides and Bifidobacterium in all weeks.
Related collections
Most cited references28
- Record: found
- Abstract: found
- Article: not found
A microbial symbiosis factor prevents intestinal inflammatory disease.
- Record: found
- Abstract: found
- Article: not found
Microbes inside--from diversity to function: the case of Akkermansia.

- Record: found
- Abstract: found
- Article: found
