- Record: found
- Abstract: found
- Article: found
Evolution of nasal and olfactory infection characteristics of SARS-CoV-2 variants
Read this article at
Abstract
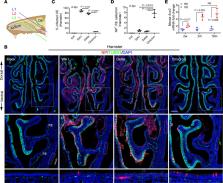
SARS-CoV-2 infection of the upper airway and the subsequent immune response are early, critical factors in COVID-19 pathogenesis. By studying infection of human biopsies in vitro and in a hamster model in vivo, we demonstrated a transition in nasal tropism from olfactory to respiratory epithelium as the virus evolved. Analyzing each variant revealed that SARS-CoV-2 WA1 or Delta infect a proportion of olfactory neurons in addition to the primary target sustentacular cells. The Delta variant possessed broader cellular invasion capacity into the submucosa, while Omicron displayed enhanced nasal respiratory infection and longer retention in the sinonasal epithelium. The olfactory neuronal infection by WA1 and the subsequent olfactory bulb transport via axon were more pronounced in younger hosts. In addition, the observed viral clearance delay and phagocytic dysfunction in aged olfactory mucosa were accompanied by a decline of phagocytosis-related genes. Further, robust basal stem cell activation contributed to neuroepithelial regeneration and restored ACE2 expression postinfection. Together, our study characterized the nasal tropism of SARS-CoV-2 strains, immune clearance, and regeneration after infection. The shifting characteristics of viral infection at the airway portal provide insight into the variability of COVID-19 clinical features, particularly long COVID, and may suggest differing strategies for early local intervention.
Abstract

Abstract
Tropism of SARS-CoV-2 in the nasal cavity has transitioned from olfactory to respiratory epithelium as the virus evolved.
Related collections
Most cited references85

- Record: found
- Abstract: found
- Article: found
A pneumonia outbreak associated with a new coronavirus of probable bat origin
- Record: found
- Abstract: found
- Article: not found