- Record: found
- Abstract: found
- Article: found
STITCH, Physicochemical, ADMET, and In Silico Analysis of Selected Mikania Constituents as Anti-Inflammatory Agents
Read this article at
Abstract
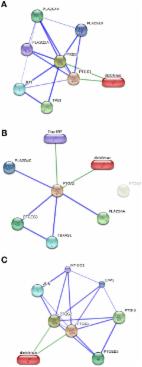
The Mikania genus has been known to possess numerous pharmacological activities. In the present study, we aimed to evaluate the interaction of 26 selected constituents of Mikania species with (i) cyclooxygenase 2 (COX 2), (ii) human neutrophil elastase (HNE), (iii) lipoxygenase (LOX), matrix metalloproteinase ((iv) MMP 2 and (v) MMP 9), and (vi) microsomal prostaglandin E synthase 2 (mPGES 2) inhibitors using an in silico approach. The 26 selected constituents of Mikania species, namely mikamicranolide, kaurenoic acid, stigmasterol, grandifloric acid, kaurenol, spathulenol, caryophyllene oxide, syringaldehyde, dihydrocoumarin, o-coumaric acid, taraxerol, melilotoside, patuletin, methyl-3,5-di-O-caffeoyl quinate, 3,3′,5-trihydroxy-4′,6,7-trimethoxyflavone, psoralen, curcumene, herniarin, 2,6-dimethoxy quinone, bicyclogermacrene, α-bisabolol, γ-elemene, provincialin, dehydrocostus lactone, mikanin-3-O-sulfate, and nepetin, were assessed based on the docking action with COX 2, HNE, LOX, MMP 2, MMP 9, and mPGES 2 using Discovery Studio (in the case of LOX, the Autodock method was utilized). Moreover, STITCH (Search Tool for Interacting Chemicals), physicochemical, drug-likeness, and ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) analyses were conducted utilizing the STITCH web server, the Mol-inspiration web server, and Discovery Studio, respectively. In the present study, STITCH analysis revealed only six ligands (dihydrocoumarin, patuletin, kaurenol, psoralen, curcumene, and nepetin) that showed interactions with human proteins. Physicochemical analysis showed that seventeen ligands complied well with Lipinski’s rule. ADMET analysis showed eleven ligands to possess hepatotoxic effects. Significantly, the binding free energy estimation displayed that the ligand methyl-3, 5-di-O-caffeoyl quinate revealed the highest binding energy for all the target enzymes, excluding LOX, suggesting that this may have efficacy as a non-steroidal anti-inflammatory drug (NSAID). The current study presents a better understanding of how Mikania is used as a traditional medicinal plant. Specifically, the 26 ligands of the Mikania plant are potential inhibitor against COX 2, HNE, LOX, MMP 2, MMP 9, and mPGES 2 for treatments for acute and/or chronic inflammatory diseases.
Related collections
Most cited references40
- Record: found
- Abstract: not found
- Article: not found
Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings1PII of original article: S0169-409X(96)00423-1. The article was originally published in Advanced Drug Delivery Reviews 23 (1997) 3–25.1
- Record: found
- Abstract: found
- Article: found
STITCH 5: augmenting protein–chemical interaction networks with tissue and affinity data
