- Record: found
- Abstract: found
- Article: found
Disruption of the bHLH transcription factor Abnormal Tapetum 1 causes male sterility in watermelon
Read this article at
Abstract
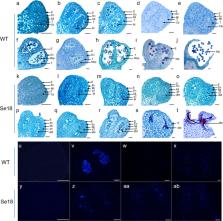
Although male sterility has been identified as a useful trait for hybrid vigor utilization and hybrid seed production, its underlying molecular mechanisms in Cucurbitaceae species are still largely unclear. Here, a spontaneous male-sterile watermelon mutant, Se18, was reported to have abnormal tapetum development, which resulted in completely aborted pollen grains. Map-based cloning demonstrated that the causal gene Citrullus lanatus Abnormal Tapetum 1 ( ClATM1) encodes a basic helix-loop-helix (bHLH) transcription factor with a 10-bp deletion and produces a truncated protein without the bHLH interaction and functional (BIF) domain in Se18 plants. qRT–PCR and RNA in situ hybridization showed that ClATM1 is specifically expressed in the tapetum layer and in microsporocytes during stages 6–8a of anther development. The genetic function of ClATM1 in regulating anther development was verified by CRISPR/Cas9-mediated mutagenesis. Moreover, ClATM1 was significantly downregulated in the Se18 mutant, displaying a clear dose effect at the transcriptional level. Subsequent dual-luciferase reporter, β-glucuronidase (GUS) activity, and yeast one-hybrid assays indicated that ClATM1 could activate its own transcriptional expression through promoter binding. Collectively, ClATM1 is the first male sterility gene cloned from watermelon, and its self-regulatory activity provides new insights into the molecular mechanism underlying anther development in plants.
Related collections
Most cited references68
- Record: found
- Abstract: found
- Article: not found
Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method.
- Record: found
- Abstract: found
- Article: not found
MEGA6: Molecular Evolutionary Genetics Analysis version 6.0.
- Record: found
- Abstract: found
- Article: not found