- Record: found
- Abstract: found
- Article: found
Physical activity is associated with slower epigenetic ageing—Findings from the Rhineland study
Read this article at
Abstract
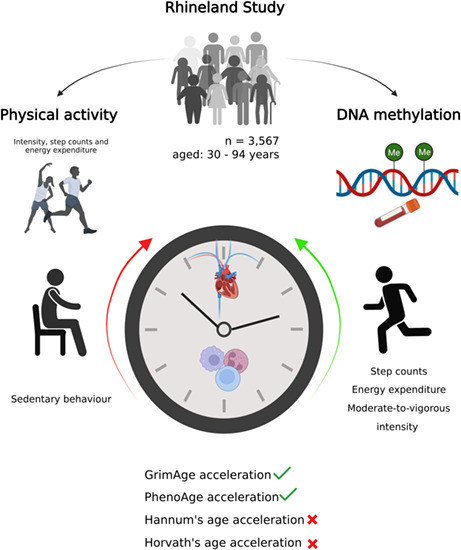
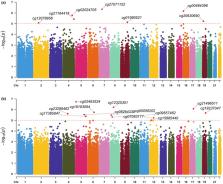
Epigenetic ageing, i.e., age‐associated changes in DNA methylation patterns, is a sensitive marker of biological ageing, a major determinant of morbidity and functional decline. We examined the association of physical activity with epigenetic ageing and the role of immune function and cardiovascular risk factors in mediating this relation. Moreover, we aimed to identify novel molecular processes underlying the association between physical activity and epigenetic ageing. We analysed cross‐sectional data from 3567 eligible participants (mean age: 55.5 years, range: 30–94 years, 54.8% women) of the Rhineland Study, a community‐based cohort study in Bonn, Germany. Physical activity components (metabolic equivalent (MET)‐Hours, step counts, sedentary, light‐intensity and moderate‐to‐vigorous intensity activities) were recorded with accelerometers. DNA methylation was measured with the Illumina HumanMethylationEPIC BeadChip. Epigenetic age acceleration (Hannum's age, Horvath's age, PhenoAge and GrimAge) was calculated based on published algorithms. The relation between physical activity and epigenetic ageing was examined with multivariable regression, while structural equation modeling was used for mediation analysis. Moreover, we conducted an epigenome‐wide association study of physical activity across 850,000 CpG sites. After adjustment for age, sex, season, education, smoking, cell proportions and batch effects, physical activity (step counts, MET‐Hours and %time spend in moderate‐to‐vigorous activities) was non‐linearly associated with slower epigenetic ageing, in part through its beneficial effects on immune function and cardiovascular health. Additionally, we identified 12 and 7 CpGs associated with MET‐Hours and %time spent in moderate‐to‐vigorous activities, respectively ( p < 1 × 10 −5). Our findings suggest that regular physical activity slows epigenetic ageing by counteracting immunosenescence and lowering cardiovascular risk.
Abstract
In this study, we examined the relation between physical activity and epigenetic ageing in 3567 eligible participants of a large community‐based, prospective cohort study, the Rhineland Study. We show that physical activity is associated with slower epigenetic ageing (GrimAge acceleration and PhenoAge acceleration), which was in part mediated by cardiovascular health and immune function.
Related collections
Most cited references53
- Record: found
- Abstract: not found
- Article: not found
lavaan: AnRPackage for Structural Equation Modeling

- Record: found
- Abstract: found
- Article: found
clusterProfiler 4.0: A universal enrichment tool for interpreting omics data
- Record: found
- Abstract: found
- Article: not found