- Record: found
- Abstract: found
- Article: found
Radiomics in pulmonary neuroendocrine tumours (NETs)
Read this article at
Abstract
Objectives
The aim of this single-centre, observational, retrospective study is to find a correlation using Radiomics between the analysis of CT texture features of primary lesion of neuroendocrine (NET) lung cancer subtypes (typical and atypical carcinoids, large and small cell neuroendocrine carcinoma), Ki-67 index and the presence of lymph nodal mediastinal metastases.
Methods
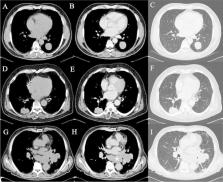
Twenty-seven patients (11 males and 16 females, aged between 48 and 81 years old—average age of 70,4 years) with histological diagnosis of pulmonary NET with known Ki-67 status and metastases who have performed pre-treatment CT in our department were included. All examinations were performed with the same CT scan (Sensation 16-slice, Siemens). The study protocol was a baseline scan followed by 70 s delay acquisition after administration of intravenous contrast medium. After segmentation of primary lesions, quantitative texture parameters of first and higher orders were extracted. Statistics nonparametric tests and linear correlation tests were conducted to evaluate the relationship between different textural characteristics and tumour subtypes.
Results
Statistically significant ( p < 0.05) differences were seen in post-contrast enhanced CT in multiple first and higher-order extracted parameters regarding the correlation with classes of Ki-67 index values. Statistical analysis for direct acquisitions was not significant. Concerning the correlation with the presence of metastases, one histogram feature (Skewness) and one feature included in the Gray-Level Co-occurrence Matrix (ClusterShade) were significant on contrast-enhanced CT only.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
Radiomics: Images Are More than Pictures, They Are Data
- Record: found
- Abstract: found
- Article: not found
Computational Radiomics System to Decode the Radiographic Phenotype
- Record: found
- Abstract: found
- Article: not found