- Record: found
- Abstract: found
- Article: found
Glomerular basement membrane deposition of collagen α1(III) in Alport glomeruli by mesangial filopodia injures podocytes via aberrant signaling through DDR1 and integrin α2β1
Read this article at
Abstract
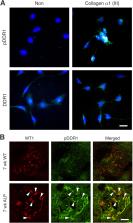
In Alport mice, activation of the endothelin A receptor (ET AR) in mesangial cells results in sub‐endothelial invasion of glomerular capillaries by mesangial filopodia. Filopodia deposit mesangial matrix in the glomerular basement membrane (GBM), including laminin 211 which activates NF‐κB, resulting in induction of inflammatory cytokines. Herein we show that collagen α1(III) is also deposited in the GBM. Collagen α1(III) localized to the mesangium in wild‐type mice and was found in both the mesangium and the GBM in Alport mice. We show that collagen α1(III) activates discoidin domain receptor family, member 1 (DDR1) receptors both in vitro and in vivo. To elucidate whether collagen α1(III) might cause podocyte injury, cultured murine Alport podocytes were overlaid with recombinant collagen α1(III), or not, for 24 h and RNA was analyzed by RNA sequencing (RNA‐seq). These same cells were subjected to siRNA knockdown for integrin α2 or DDR1 and the RNA was analyzed by RNA‐seq. Results were validated in vivo using RNA‐seq from RNA isolated from wild‐type and Alport mouse glomeruli. Numerous genes associated with podocyte injury were up‐ or down‐regulated in both Alport glomeruli and cultured podocytes treated with collagen α1(III), 18 of which have been associated previously with podocyte injury or glomerulonephritis. The data indicate α2β1 integrin/DDR1 co‐receptor signaling as the dominant regulatory mechanism. This may explain earlier studies where deletion of either DDR1 or α2β1 integrin in Alport mice ameliorates renal pathology. © 2022 Boys Town National Research Hospital. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.
Related collections
Most cited references51

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data
- Record: found
- Abstract: found
- Article: not found
STAR: ultrafast universal RNA-seq aligner.
- Record: found
- Abstract: found
- Article: not found