- Record: found
- Abstract: found
- Article: found
Spatially patterned excitatory neuron subtypes and projections of the claustrum
Read this article at
Abstract
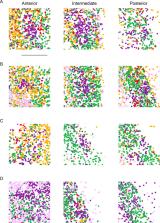
The claustrum is a functionally and structurally complex brain region, whose very spatial extent remains debated. Histochemical-based approaches typically treat the claustrum as a relatively narrow anatomical region that primarily projects to the neocortex, whereas circuit-based approaches can suggest a broader claustrum region containing projections to the neocortex and other regions. Here, in the mouse, we took a bottom-up and cell-type-specific approach to complement and possibly unite these seemingly disparate conclusions. Using single-cell RNA-sequencing, we found that the claustrum comprises two excitatory neuron subtypes that are differentiable from the surrounding cortex. Multicolor retrograde tracing in conjunction with 12-channel multiplexed in situ hybridization revealed a core-shell spatial arrangement of these subtypes, as well as differential downstream targets. Thus, the claustrum comprises excitatory neuron subtypes with distinct molecular and projection properties, whose spatial patterns reflect the narrower and broader claustral extents debated in previous research. This subtype-specific heterogeneity likely shapes the functional complexity of the claustrum.
Related collections
Most cited references56
- Record: found
- Abstract: found
- Article: not found
Fiji: an open-source platform for biological-image analysis.
- Record: found
- Abstract: found
- Article: not found
STAR: ultrafast universal RNA-seq aligner.
- Record: found
- Abstract: found
- Article: not found