- Record: found
- Abstract: found
- Article: found
Reversible Signal Binding by the Pseudomonas aeruginosa Quorum-Sensing Signal Receptor LasR
Read this article at
Abstract
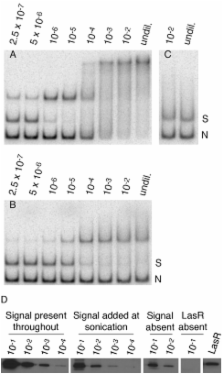
Many members of the LuxR family of acyl-homoserine lactone (acyl-HSL)-dependent quorum-sensing transcriptional activators are thought to have the unusual characteristics of requiring the signal ligand during polypeptide synthesis to fold into an active conformation and of binding signal extraordinarily tightly. This is the case for the N-3-oxo-dodecanoyl-HSL-dependent Pseudomonas aeruginosa virulence regulator LasR. We present evidence that LasR can fold into an active conformation in vivo in the absence of the acyl-HSL ligand. We also present evidence indicating that in the cellular environment, LasR and N-3-oxo-dodecanoyl-HSL readily dissociate. After dissociation, LasR can remain in a properly folded conformation capable of reassociating with signal. We present a new model for the folding and signal binding of LasR and other members of the family of transcription factors to which LasR belongs. Our findings have important implications concerning the cellular responses to decreased environmental concentrations of signals and have implications about potential quorum-sensing inhibition strategies.
IMPORTANCE
The opportunistic pathogen Pseudomonas aeruginosa causes difficult to treat and incurable diseases. Quorum sensing controls expression of many virulence factors in this bacterium, and the quorum-sensing signal receptor LasR has been targeted for anti-quorum-sensing therapeutic development. The work described here changes the current view about how LasR interacts with the quorum-sensing signal to which it responds. This is of importance for therapeutic development. The experiments also address a poorly studied aspect of quorum sensing: the response to decreases in quorum-sensing signals. This has importance with respect to understanding the ecology and evolution of quorum sensing.
Related collections
Most cited references32
- Record: found
- Abstract: found
- Article: not found
Quorum sensing: cell-to-cell communication in bacteria.
- Record: found
- Abstract: found
- Article: not found
Listening in on bacteria: acyl-homoserine lactone signalling.
- Record: found
- Abstract: found
- Article: not found